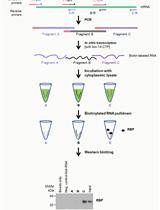
Protocols for Coronavirus/COVID-19 Research
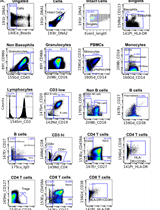
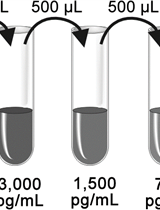
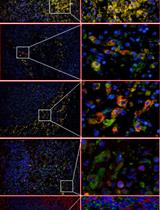
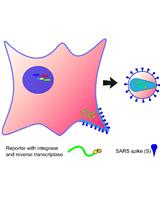
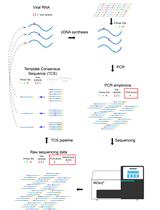
With the spread of the COVID-19 (SARS-CoV-2) pandemic across the world, sharing accurate and reproducible methods that can be readily available to the biomedical community became crucial. Therefore, Bio-protocol decided to dedicate a special issue to protocols used in Coronavirus/COVID-19 research in May 2020. By November 2021, we have published 19 high quality protocols used in basic science and clinical research, including diagnostic methods for COVID-19. Additionally, 9 protocols published before May 2020, used in tangential research on other coronaviruses, are also included in this special issue.
More new protocols will be welcomed to be included in this special issue, and we hope it will become a comprehensive resource for detailed protocols relevant to Coronavirus/COVID-19 research. Typically, we only accept protocols previously used in published articles reporting original research. However, due to the rapid rate of preprint publications during the pandemic, we also include protocols used in preprint articles and preprint versions of the protocols.
















































 (3).jpg)