- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Functional Evaluation of the Signal Peptides of Secreted Proteins
Published: Vol 8, Iss 9, May 5, 2018 DOI: 10.21769/BioProtoc.2839 Views: 12545
Reviewed by: Xiaolin ChenAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Evaluation of Translation Rate Through L-azidohomoalanine (AHA) Incorporation and Subsequent Alkyne Fluorophore–Mediated Click Chemistry in Yeast
Mainak Pratim Jha and Koyeli Mapa
Jul 20, 2025 2544 Views

In Vitro Screening of Microbial Extracts Against the Oomycetes Phytophthora capsici and Pythium ultimum
Mónica Trigal Martínez [...] María Ángeles Vinuesa Navarro
Sep 20, 2025 1261 Views

A Reliable In Planta Inoculation and Antifungal Screening Protocol for Rhizoctonia solani-Induced Sheath Blight in Rice
Alinaj Yasin [...] Palash Deb Nath
Nov 5, 2025 1568 Views
Abstract
Here, we describe a method that can be used to evaluate the function of predicted signal peptides. This method utilizes the yeast Saccharomyces cerevisiae YTK12 strain and pSUC2 vector in which the pSUC2 vector with fused predicted signal peptide is transformed into yeast. The function of the signal peptides can be evaluated by using different selective media and color reaction. In this protocol, we provide the detailed description of manipulation in order to implement easily.
Keywords: Secreted proteinBackground
Microbial eukaryotes, such as fungi and oomycetes, secret abundant of proteins to play a variety of functions. Currently, the most widely used method for prediction of signal peptide from amino acid sequences is to use software SignalP (Petersen et al., 2011). The yeast YTK12 strain is invertase negative and pSUC2 vector contains invertase gene but lack Methionine (Met) and signal peptide sequence, thus YTK12 strain and pSUC2 vector were widely used for biological evaluation of peptide secretion (Jacobs et al., 1997; Oh et al., 2009). In addition, the color reaction could be used to verify the result, because the invertase enzymatic activity can be detected by the reduction of 2,3,5-Triphenyltetrazolium Chloride (TTC) to insoluble red colored 1,3,5-Triphenylformazan (TPF). Although the method has been generally used to evaluate the function of signal peptides, there is no specific and detailed information about this method (Oh et al., 2009; Song et al., 2015). Here, we describe a modified high-efficiency yeast transformation method (Gietz and Schiestl, 2007) and detailed protocol to evaluate the function of signal peptides, which will be the primary part for the secretory proteins research.
Materials and Reagents
- Pipette tips
- 1.5 ml tubes
- 2 ml tubes
- 10 ml test tube
- Millipore filter units, 0.22 µm (Millex-GP, Merck, catalog number: SLGP033RB )
- Yeast YTK12 strain
- pSUC2 vector
- EcoRI restriction enzymes (Takara Bio, catalog number: 1040S )
- XhoI restriction enzymes (Takara Bio, catalog number: 1094S )
- Yeast extract (OXOID, catalog number: LP0021 )
- Peptone (BD, DifcoTM, catalog number: 211677 )
- Glucose (Sinopharm Chemical Reagent, catalog number: 10010518 )
- Agar A (Sangon Biotech, catalog number: A600010 )
- Adenine hemisulfate salt (Sigma-Aldrich, Vetec, catalog number: V900375 )
- Tris-HCl (Sigma-Aldrich, catalog number: V900312 )
- EDTA (Sinopharm Chemical Reagent, catalog number: 10009617 )
- NaOH (Sinopharm Chemical Reagent, catalog number: 10019718 )
- Single-stranded carrier DNA (salmon sperm DNA, Solarbio, catalog number: D8030 )
- LiAc (Sinopharm Chemical Reagent, catalog number: 30109760 )
- PEG (Polyethylene glycol, Sigma-Aldrich, catalog number: P3640 )
- DMSO (Dimethyl Sulfoxide, Sigma-Aldrich, catalog number: D5879 )
- YNB (yeast nitrogen base without amino acids, BD, DifcoTM, catalog number: 291920 )
- -Trp DO supplement (Clontech, catalog number: 630413 )
- Sucrose (Sinopharm Chemical Reagent, catalog number: 10021418 )
- Antimycin A (Sigma-Aldrich, catalog number: A8674 )
- Raffinose (D-(+)-Raffinose pentahydrate, Bomei, CAS Number: 17629-30-0)
- Sodium acetate (Sinopharm Chemical Reagent, catalog number: 10018718 )
- Acetic acid (Sinopharm Chemical Reagent, catalog number: 10000218 )
- KH2PO4 (Sinopharm Chemical Reagent, catalog number: 10017618 )
- Na2HPO4 (Sinopharm Chemical Reagent, catalog number: 10020318 )
- TTC (Tokyo Chemical Industry (TCI), CAS Number: 298-96-4)
- YPD medium (1 L) (see Recipes)
- 1x TE Buffer (100 ml) (see Recipes)
- Single-stranded carrier DNA (2 mg/ml) (see Recipes)
- 1.0 M LiAc (100 ml) (see Recipes)
- 50% (w/v) PEG (100 ml) (see Recipes)
- CMD-W medium (1 L) (see Recipes)
- Antimycin A stock solution (50 mg/ml) (see Recipes)
- YPRAA medium (1 L) (see Recipes)
- 10 mM acetic acid-sodium acetate buffer (100 ml, pH = 4.7) (see Recipes)
- Phosphate Buffer (150 ml) (see Recipes)
- TTC Stock solution (2%) (see Recipes)
Equipment
- Pipettes (Mettler-Toledo International, RAININ, model: XLS )
- Incubator (ZHICHENG, model: ZXDP-B2050 )
- Clean bench (AIRTECH, model: SW-CJ-2FD , catalog number: A11062689)
- Water bath (Shanghai Jinghong Laboratory Instrument, model: DK-8D )
- Vortex mixer (Kylin-Bell Lab Instruments, model: VORTEX-5 )
- Incubator shaker (Changzhou Zhiborui Instrument Manufacturing, model: THZ-D )
- Centrifuge (Eppendorf, model: 5424 )
Procedure
- pSUC2 vector construction
- DNA fragments that code the predicted signal peptides and the following additional two amino acids were amplified and introduced into pSUC2 using EcoRI and XhoI restriction sites (Figure 1). The DNA sequences around enzyme cutting site are as follows and the sequences of sites are underlined.
EcoRI: 5’ TCCAAGCTCGGAATTTTAATTAAGAATTC 3’
XhoI: 5’ CTCGAGGTTCTCCCTATAGTGAGTCGTAT 3’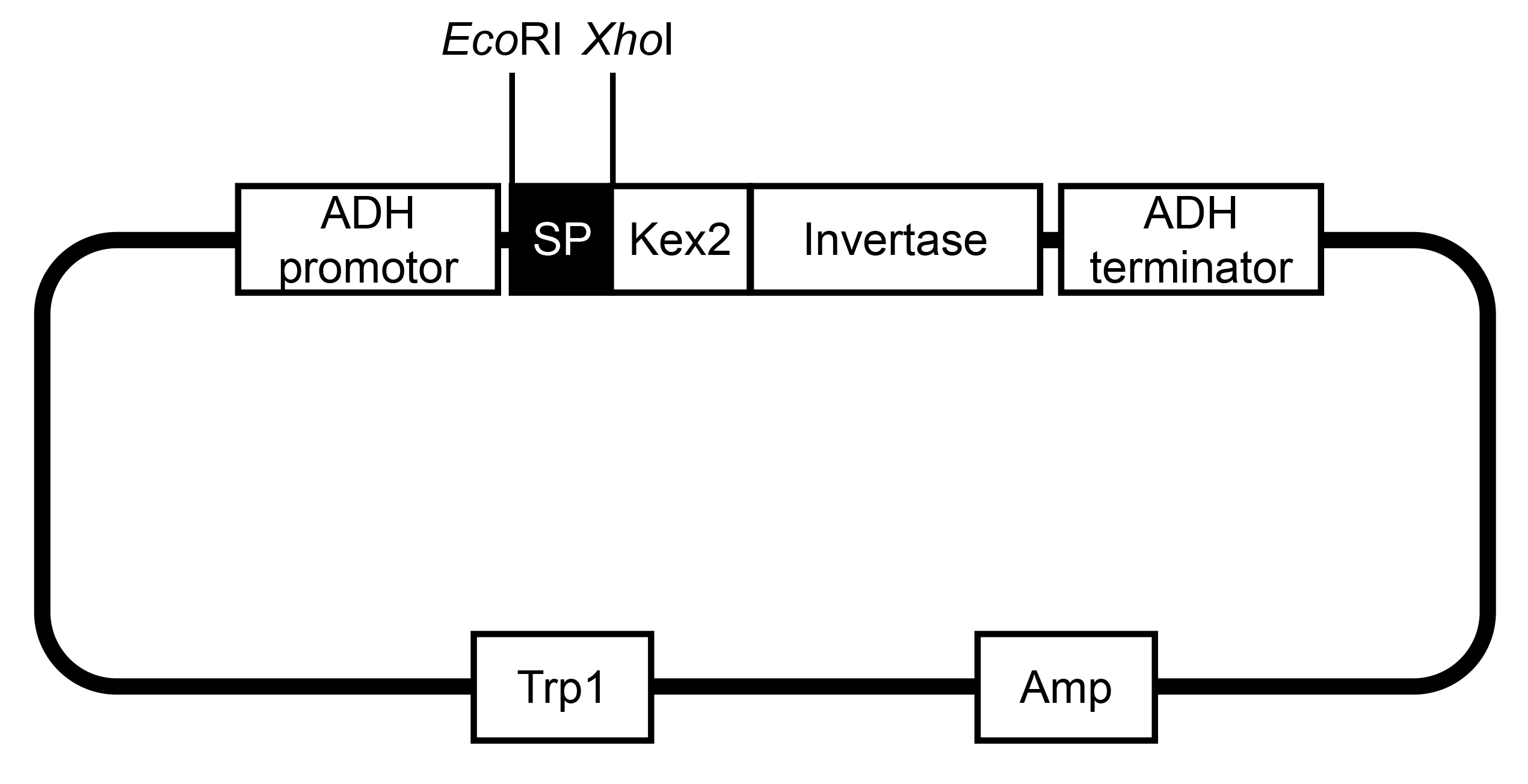
Figure 1. The vector information of pSUC2. This map was modified from Jacobs (Jacobs et al., 1997). - Confirm the positive transformants by PCR with pSUC2 vector-specific primers as follows, which should be further confirmed by sequencing.
pSUC2F: GGTGTGAAGTGGACCAAAGGTCTA
pSUC2R: CCTCGTCATTGTTCTCGTTCCCTT - In this study, we use the recombinant plasmid that contains Avr1b signal peptide from Phytophthora sojae that was demonstrated to be functional (Dou et al., 2008; Song et al., 2015).
- DNA fragments that code the predicted signal peptides and the following additional two amino acids were amplified and introduced into pSUC2 using EcoRI and XhoI restriction sites (Figure 1). The DNA sequences around enzyme cutting site are as follows and the sequences of sites are underlined.
- Transformation of plasmid DNA into yeast cells
- Streak an YPDA agar plate with the YTK12 yeast cells from a frozen yeast stock, incubate the plate upside down at 30 °C for 2-3 days until colonies appear.
Note: YPD medium also can be used, although yeast can get a better situation on YPDA. If there are no special instructions, the following steps are done at the room temperature. - Inoculate one colony (diameter 2-3 mm) into 3 ml YPDA liquid medium in a sterile test tube, incubate at 30 °C for 16-18 h with shaking at 220 rpm and measure the OD values at 600 nm to get suspension with OD value more than 0.6 (OD600 > 0.6).
- Pipette about 1.5 ml of cell suspension into a 2 ml tube and centrifuge the cells at 12,000 x g for 1 min at room temperature.
- Discard the supernatant and re-suspend the cells with 1.5 ml sterile distilled water or TE buffer.
- Centrifuge the cells at 12,000 x g for 1 min and discard the supernatant.
- For each transformation, add reagents in a 2 ml tube as following order and keep on ice: 34 μl plasmid DNA and sterile water (1 μg Plasmid DNA will be enough for transformation); 50 μl single-stranded carrier DNA (2.0 mg/ml) and mix well; 36 μl 1.0 M LiAc and mix well; 240 μl 50% (w/v) PEG 3350 and vortex mix vigorously or use a sterile pipette tip to break up the cell pellet to mix thoroughly.
- Incubate with shaking at 220 rpm for 30 min at 30 °C.
- Add 1/10 of the volume DMSO and mix gently.
- Incubate the cell suspension at 42 °C for 15 min and mix upside down for several times during this time.
- Incubate on ice for 2 min.
- Centrifuge at 12,000 x g for 1 min and discard the supernatant.
- Add 100 μl sterile distilled water or TE buffer and re-suspend the cells with a pipette carefully.
- Coat the cell suspension on selective media CMD-W plate using a sterile glass rod and incubate at 30 °C for 3 days.
- Streak an YPDA agar plate with the YTK12 yeast cells from a frozen yeast stock, incubate the plate upside down at 30 °C for 2-3 days until colonies appear.
- Screening the positive yeast colonies on selective media
- After transformation, spread the yeast cells on CMD-W media plates.
- Streak the colonies to fresh CMD-W plates and incubate at 30 °C for at least 2 days.
- Streak the colonies to new CMD-W plates and YPRAA plates (raffinose media) and incubate for another 2-3 days at 30 °C.
- After transformation, spread the yeast cells on CMD-W media plates.
- Color observation to detect signal peptide secretion function
- The color change is used to detect signal peptide secretion function.
- Streak the single transformational colony into CMD-W liquid medium, the YTK12 yeast that does not contain any plasmid DNA was cultured in YPDA liquid medium as control.
- Incubate with shaking at 220 rpm for 36 h at 30 °C.
- Pipette about 1.5 ml of cell suspension into a 2 ml tube and centrifuge at 12,000 x g for 1 min at room temperature.
- Discard the supernatant, add 1.5 ml sterile distilled water and vortex shock to mix vigorously.
- Centrifuge at 12,000 x g for 1 min, discard the supernatant and add 1.5 ml sterile distilled water and vortex shock to mix well.
- Centrifuge at 12,000 x g for 1 min and discard the supernatant.
- Re-suspend with 750 μl sterile distilled water, add 250 μl 10 mM acetic acid–sodium acetate buffer (pH = 4.7), 500 μl 10 % sucrose solution (w/v), incubate at 37 °C for 10 min.
- Centrifuge at 12,000 x g for 1 min, take 100 μl of the supernatant, put into glass test tube and add 900 μl 0.1% TTC solution, incubate at room temperature for 5 min. Colorimetric change will be observed if the signal peptide is functional, while the red color will be not observed for YTK12 strain and YTK12 carrying the empty pSUC2 vector.
Note: The empty pSUC2 vector also contains a non-functional sequence.
- The color change is used to detect signal peptide secretion function.
Data analysis
If the signal peptide is functional, the yeast transformants can grow on CMD-W and YPRAA medium plates and the solution will turn red in the color test experiment (Figure 2).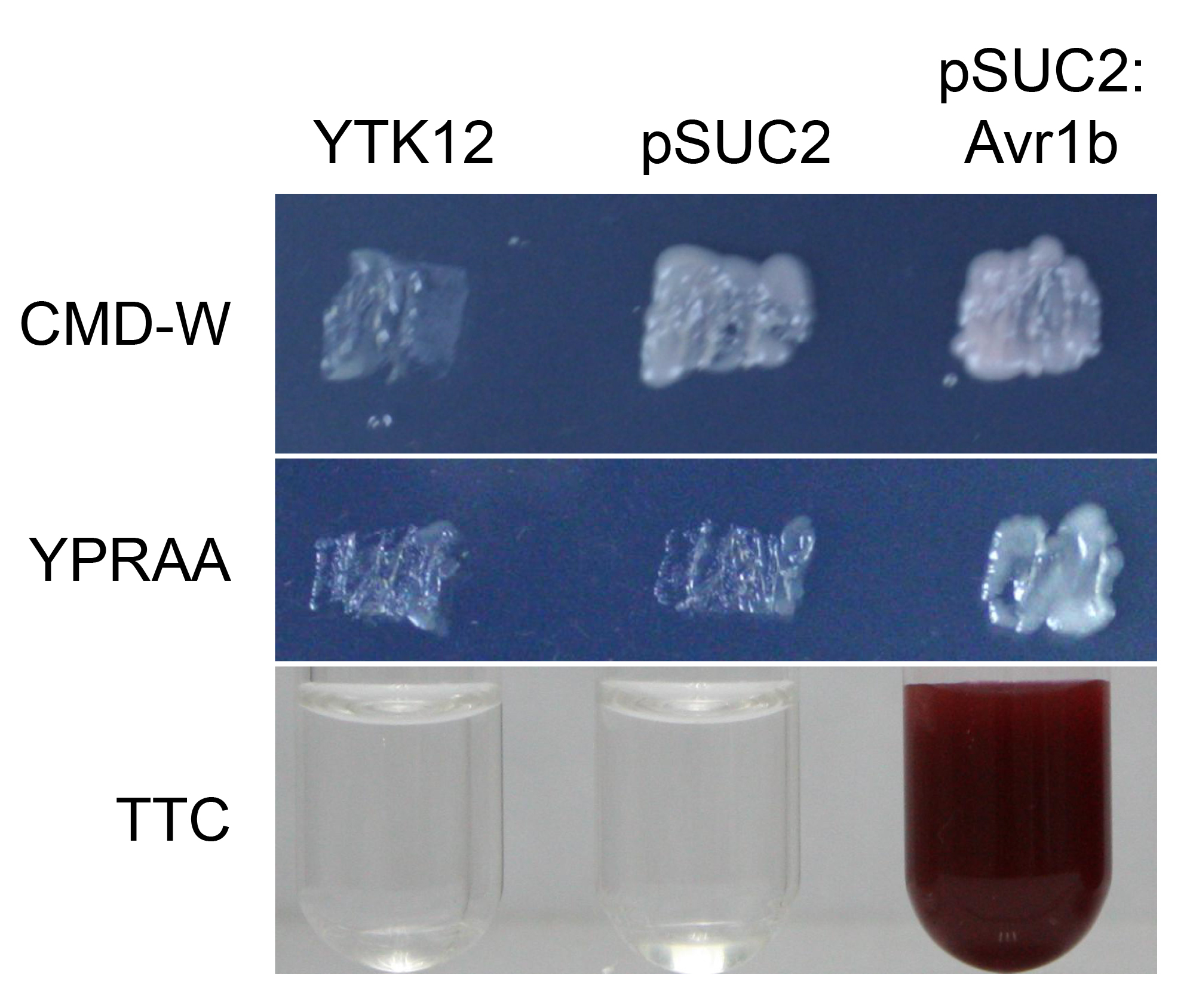
Figure 2. Functional validation of the signal peptide of Avr1b.The yeast YTK12 strain carrying Avr1b signal peptide fragment fused in the pSUC2 vector are able to grow in both the CMD-W and YPRAA media, and also induce red color reaction, indicating the secretory function. The YTK12 strain and YTK12 carrying the empty pSUC2 vector served as controls.
Notes
Inoculate colony on CMD-W plates is to verify if the plasmid DNA is transformed into the yeast. We also observed that the yeast without any vector can get a small amount of growth on CMD-W and YPRAA plates under the condition of incubation for a long time (more than three days). To assay the invertase secretion, colonies were plated on YPRAA plates containing raffinose but lacking glucose for several times. The transformants containing pSUC2 vector can grow on YPRAA plate if incubated for more than three days. Thus, when the yeast colonies are streaked on CMD-W and YPRAA plates, it must be observed every day. The principle of color change is that the invertase enzymatic activity was detected by the reduction of 2,3,5-Triphenyltetrazolium Chloride (TTC) to insoluble red colored 1,3,5-Triphenylformazan (TPF).
Recipes
- YPD medium (1 L)
10 g yeast extract
20 g peptone
20 g glucose
Add distilled water to 1 L, autoclave for 15 min at 121 °C
Add 20 g Agar A for solid medium
Supplement of adenine hemisulfate to the final concentration 30 mg/L in YPD could provide yeast a better growth condition - 1x TE Buffer (100 ml)
1 M Tris-HCl (pH 7.5) 10 ml
500 mM EDTA (pH 8.0) 2 ml
Adjust pH to 7.5 with NaOH
Make up to 100 ml
Autoclave for 15 min at 121 °C - Single-stranded carrier DNA (2 mg/ml)
- Dissolve 100 mg single-stranded carrier DNA in 50 ml of sterile TE buffer by using magnetic stir at 4 °C until no visible DNA is seen
- Dispense 1.0 ml sample into 1.5 ml tubes and denature the carrier DNA in a boiling water bath for 5 min and chill immediately in an ice/water bath for several minutes, and store at -20 °C
- Make sure to gradually thaw the denatured carrier DNA on ice whenever it is used
- Denatured carrier DNA could be boiled, frozen and thawed many times
- Dissolve 100 mg single-stranded carrier DNA in 50 ml of sterile TE buffer by using magnetic stir at 4 °C until no visible DNA is seen
- 1.0 M LiAc (100 ml)
10.2 g LiAc
Add distilled water to the volume of 100 ml
Sterilize by filtering Millipore filter units, 0.22 µm - 50% (w/v) PEG (100 ml)
50 g PEG
Add 80 ml distilled water to fully dissolved and then make the volume up to 100 ml
Autoclave for 15 min at 121 °C - CMD-W medium (1 L)
6.7 g YNB (yeast nitrogen base without amino acids)
0.74 g -Trp DO supplement
20 g sucrose
1 g glucose
20 g Agar A
Add distilled water to 1 L, autoclave for 15 min at 121 °C - Antimycin A stock solution (50 mg/ml)
50 mg antimycin A dissolved in DMSO (dimethylsulphoxide) - YPRAA medium (1 L)
10 g yeast extract
20 g peptone
20 g raffinose
20 g Agar A
Add distilled water to 1 L, autoclave for 15 min at 121 °C
Add antimycin A to a final concentration of 2 mg/ml just before pouring medium plates - 10 mM acetic acid-sodium acetate buffer (100 ml, pH = 4.6)
0.136 g sodium acetate
Add 90 ml water
Adjust pH to 7.5 with acetic acid
Make up to 100 ml - Phosphate Buffer (150 ml)
100 ml 66.7 mM KH2PO4
50 ml 68.6 mM
Mix well to get phosphate buffer - TTC Stock solution (2%)
- Dissolve 2 g TTC in 100 ml water, store in brown bottle or keep away from light
Note: Because TTC in water cannot be kept for a long time, it should be dissolved in phosphate buffer for long-term preservation. - Dilute with 1 M NaOH to 0.1% when it is used
- Dissolve 2 g TTC in 100 ml water, store in brown bottle or keep away from light
Acknowledgments
This research was supported by the National Natural Science Foundation of China (Grant No. 31701736) and the Fundamental Research Funds for the Central Universities (Grant Nos. 2662015PY195 and 2662017JC003). The authors have declared that no conflicts of interest or competing interests exist.
References
- Dou, D., Kale, S. D., Wang, X., Jiang, R. H., Bruce, N. A., Arredondo, F. D., Zhang, X. and Tyler, B. M. (2008). RXLR-mediated entry of Phytophthora sojae effector Avr1b into soybean cells does not require pathogen-encoded machinery. Plant Cell 20(7): 1930-1947.
- Gietz, R. D. and Schiestl, R. H. (2007). Large-scale high-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat Protoc 2(1): 38-41.
- Jacobs, K. A., Collins-Racie, L. A., Colbert, M., Duckett, M., Golden-Fleet, M., Kelleher, K., Kriz, R., LaVallie, E. R., Merberg, D., Spaulding, V., Stover, J., Williamson, M. J. and McCoy, J. M. (1997). A genetic selection for isolating cDNAs encoding secreted proteins. Gene 198(1-2): 289-296.
- Oh, S. K., Young, C., Lee, M., Oliva, R., Bozkurt, T. O., Cano, L. M., Win, J., Bos, J. I., Liu, H. Y., van Damme, M., Morgan, W., Choi, D., Van der Vossen, E. A., Vleeshouwers, V. G. and Kamoun, S. (2009). In planta expression screens of Phytophthora infestans RXLR effectors reveal diverse phenotypes, including activation of the Solanum bulbocastanum disease resistance protein Rpi-blb2. Plant Cell 21(9): 2928-2947.
- Petersen, T. N., Brunak, S., von Heijne, G. and Nielsen, H. (2011). SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8(10): 785-786.
- Song, T., Ma, Z., Shen, D., Li, Q., Li, W., Su, L., Ye, T., Zhang, M., Wang, Y. and Dou, D. (2015). An oomycete CRN effector reprograms expression of plant HSP genes by targeting their promoters. PLoS Pathog 11(12): e1005348.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Yin, W., Wang, Y., Chen, T., Lin, Y. and Luo, C. (2018). Functional Evaluation of the Signal Peptides of Secreted Proteins. Bio-protocol 8(9): e2839. DOI: 10.21769/BioProtoc.2839.
Category
Plant Science > Plant immunity > Host-microbe interactions
Molecular Biology > Protein > Detection
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










