- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Isolation of Epithelial and Stromal Cells from Colon Tissues in Homeostasis and Under Inflammatory Conditions
(*contributed equally to this work, § Technical contact) Published: Vol 13, Iss 18, Sep 20, 2023 DOI: 10.21769/BioProtoc.4825 Views: 5638
Reviewed by: Kazem NouriArundhati MehtaAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Single Cell Isolation from Human Diabetic Fibrovascular Membranes for Single-Cell RNA Sequencing
Katia Corano Scheri [...] Amani A. Fawzi
Oct 20, 2024 1885 Views

RACE-Nano-Seq: Profiling Transcriptome Diversity of a Genomic Locus
Lu Tang [...] Philipp Kapranov
Jul 5, 2025 2504 Views
Analyzing RNA Localization Using the RNA Proximity Labeling Method OINC-seq
Megan C. Pockalny [...] J. Matthew Taliaferro
Aug 5, 2025 2343 Views
Abstract
Inflammation of the gastrointestinal tract is a prevalent pathology in diseases such as inflammatory bowel disease (IBD). Currently, there are no therapies to prevent IBD, and available therapies to treat IBD are often sub-optimal. Thus, an unmet need exists to better understand the molecular mechanisms underlying intestinal tissue responses to damage and regeneration. The recent development of single-cell RNA (sc-RNA) sequencing-based techniques offers a unique opportunity to shed light on novel signaling pathways and cellular states that govern tissue adaptation or maladaptation across a broad spectrum of diseases. These approaches require the isolation of high-quality cells from tissues for downstream transcriptomic analyses. In the context of intestinal biology, there is a lack of protocols that ensure the isolation of epithelial and non-epithelial compartments simultaneously with high-quality yield. Here, we report two protocols for the isolation of epithelial and stromal cells from mouse and human colon tissues under inflammatory conditions. Specifically, we tested the feasibility of the protocols in a mouse model of dextran sodium sulfate (DSS)-induced colitis and in human biopsies from Crohn’s patients. We performed sc-RNA sequencing analysis and demonstrated that the protocol preserves most of the epithelial and stromal cell types found in the colon. Moreover, the protocol is suitable for immunofluorescence staining of surface markers for epithelial, stromal, and immune cell lineages for flow cytometry analyses. This optimized protocol will provide a new resource for scientists to study complex tissues such as the colon in the context of tissue damage and regeneration.
Key features
• This protocol allows the isolation of epithelial and stromal cells from colon tissues.
• The protocol has been optimized for tissues under inflammatory conditions with compromised cell viability.
• This protocol is suitable for experimental mouse models of colon inflammation and human biopsies.
Graphical overview
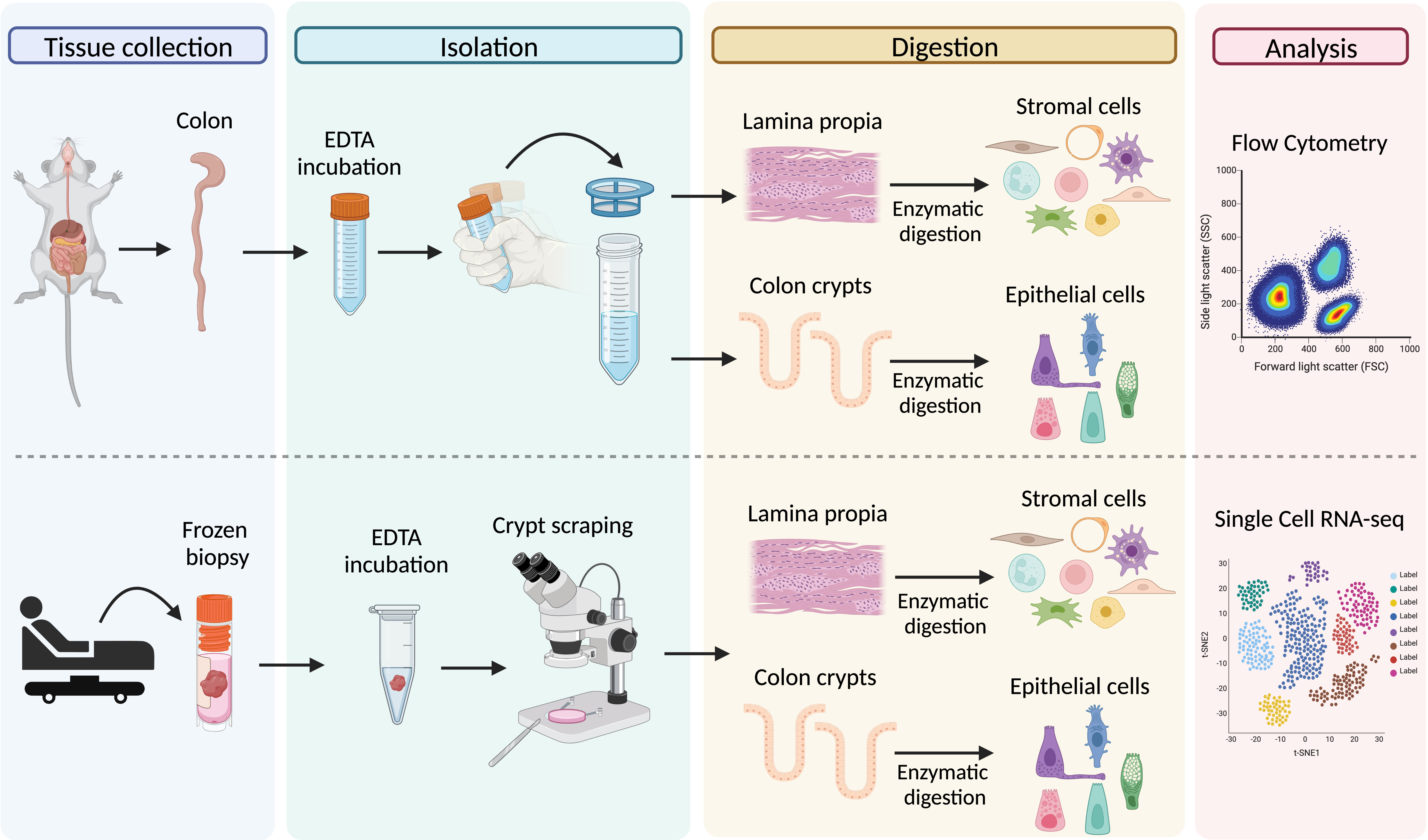
Graphical representation of the main steps for the processing of colon tissue from dextran sodium sulfate (DSS)-treated mice (upper panel) and frozen biopsies from Crohn’s patients (lower panel)
Background
Inflammatory bowel disease (IBD) is a chronic inflammatory condition of the gastrointestinal (GI) tract that encompasses two distinct pathologies: ulcerative colitis and Crohn’s disease. The global prevalence of IBD has been increasing since 2000 and now affects approximately 1.4 million individuals in America and 2.3 million in Europe (Ng et al., 2017). Current medical treatment for IBD involves the use of immunomodulatory agents to empirically dampen inflammation; however, up to one-third of patients fail initial treatment, and half of the initial responders will eventually relapse.
The interplay between the epithelial and stromal compartments supports homeostasis of the GI tract. Epithelial cells form a dynamic cellular layer with high regeneration potential that separates the luminal content from the underlying stromal tissue while supporting nutrient and water absorption. Simultaneously, the resident stromal cells of the lamina propria provide structural support, immunity against pathogens, and tolerance to the commensal microbiota. Breakdown of this equilibrium by dysregulation of host–microbiome interactions, defects in intestinal epithelial barrier permeability, and loss of immune tolerance can lead to the development of IBD (Maloy and Powrie, 2011). Therefore, it is important to consider the complex cellular heterogeneity when designing new experimental approaches to better understand the molecular mechanisms underlying disease.
The recent development of single-cell technologies has provided a finer picture of complex biology and deciphered heterogeneity present in tissue, enabling the identification of new cell populations, cellular interactions, and signaling pathways associated with pathology. However, these powerful approaches require the isolation of high-quality single cells from tissues to provide a representative and reliable tissue-associated cellular landscape. While the field of single-cell technology is moving fast, scientists have had to adapt tissue dissociation protocols depending on the cell types and contexts of interest. In the context of intestinal biology, the complex cellular heterogeneity of the gut mucosa and sensitivity to detachment-mediated cell death has hindered protocols for tissue dissociation that aim to represent the cellular repertoire of the original tissue. The main limitation is the fragility of the epithelial cells that cannot support most enzymes used to dissociate tissues. This becomes even more challenging when working with tissues under inflammatory conditions where cells have been exposed to a harmful and often cytotoxic environment. Some studies have already developed protocols to obtain epithelial and stromal cells from the same tissues (Smillie et al., 2019; Elmentaite et al., 2020); however, there is variability across different studies and laboratories. Our goal was to establish a detailed and easy-to-follow protocol to isolate simultaneously epithelial and stromal cells from mouse and human colon samples under inflammatory conditions. For the mouse protocol, we used the common dextran sodium sulfate (DSS) model of intestinal damage; for the human, we optimized the protocol for processing cryo-preserved colon biopsies from patients with Crohn’s disease. We validated the protocols using downstream flow cytometry and single-cell RNA sequencing analyses and demonstrated that both recover the main cell populations found in the gut mucosa.
Protocol for processing colon tissues from DSS-treated mice
Materials and reagents
Biological materials
7-week-old female CL57BL/6 mice (Charles River Laboratory) were treated with 3% dextran sodium sulfate (DSS) in water for five consecutive days. Colon tissues were isolated on day 3 after treatment at the peak of the inflammatory response.
Reagents
RPMI (Invitrogen, catalog number: 11875119)
Advanced DMEM/F-12 (Gibco, catalog number: 12-634-028)
HBSS (Cytiva, catalog number: SH3058801)
HEPES (Invitrogen, catalog number: 15630080)
GlutaMAX (Thermo Fisher Scientific, catalog number: 35050061)
Pen/Strep (Invitrogen, catalog number: 15140122)
EDTA (0.5 M, pH 8.0) (Invitrogen, catalog number: AM9260G)
DNase I (Roche, catalog number: 10104159001)
Liberase TM (Sigma, catalog number: 05401127001)
TrypLE (Invitrogen, catalog number: 12605010)
Dextran sodium sulfate (DSS) (Alfa Aesar, catalog number: J62101-22)
Trypan Blue solution (Gibco, catalog number: 15250061)
Flow cytometry antibodies:
LIVE/DEADTM Fixable Aqua Dead Cell Stain kit (Thermo Fisher Scientific, catalog number: L34957)
BV421 Rat anti-mouse CD31 (BD Horizon, catalog number: 562939, clone: MEC 13.3)
APC Anti-mouse CD326 (EP-CAM) (BioLegend, catalog number: 118214, clone: G8.8)
PE Anti-mouse Podoplanin (BioLegend, catalog number: 127408, clone: 8.1.1)
FITC Rat anti-mouse CD45 (BD Pharmingen, catalog number: 553080, clone: 30-F11)
Note: Depending on the flow cytometer available, users can use a different color panel design.
Solutions
Epithelial cell solution (see Recipes)
Lamina propria cell solution (see Recipes)
Epithelial wash buffer (see Recipes)
Recipes
Epithelial cell solution
Prepare it fresh and keep it on ice.
Reagent Final concentration Quantity (for one sample) HBSS n/a 11,250 μL HEPES 10 mM 150 μL EDTA (0.5 M) 10 mM 300 μL Pen/Strep 100 U/mL 150 μL FBS 2% 300 μL DNase I 100 μg/mL 150 μL Total n/a 15 mL Lamina propria solution
Prepare it fresh and keep it on ice.
Reagent Final concentration Quantity (for one sample) RPMI n/a 9,600 μL Pen/Strep 100 U/mL 100 μL Liberase TM 100 μg/mL 200 μL DNase I 100 μg/mL 100 μL Total n/a 10 mL Epithelial wash buffer
Once prepared can be stored at 4 °C for one month.
Reagent Final concentration Quantity (for one sample) Adv-DMEM F12 n/a 4,900 μL HEPES 10 mM 500 μL GlutaMAX 10 mM 500 μL Total n/a 50 mL
Laboratory supplies
15 mL centrifuge tube (CELLTREAT, catalog number: 30411B)
50 mL centrifuge tube (CELLTREAT, catalog number: 229430)
100 mm × 15 mm Petri dishes (CELLTREAT, catalog number: 229693)
100 μm cell strainer (Falcon, catalog number: 352360)
40 μm cell strainer (Falcon, catalog number: 352340)
Gavage needle (Gavage Needle, catalog number: AFN2025S)
Frosted microscope slides (Fisher Scientific, catalog number: 12-550-343)
5 mL Polystyrene round-bottom tube with cell-strainer cap (Falcon, catalog number: 352235)
Filter pipette tips (1,000 μL) (Genesee Scientific, catalog number: 24-430)
Serological pipets (10 and 5 mL) (Genesee Scientific, catalog number: 12-102, 12-104)
Equipment
Surgical scissors
Forceps
Centrifuge 5810 R (Eppendorf)
Countess 3 (Invitrogen)
Water bath (Fisher Scientific, Isotemp GPD 10)
Software and datasets
FlowJo v10.8.1
GraphPad Prism9
Procedure
Harvesting colon tissue and cleaning
Cut the colon from the gastrointestinal tract.
Note: Colons from DSS-treated mice tend to be shorter compared to control mice, as a result of active inflammation (Figure 1).
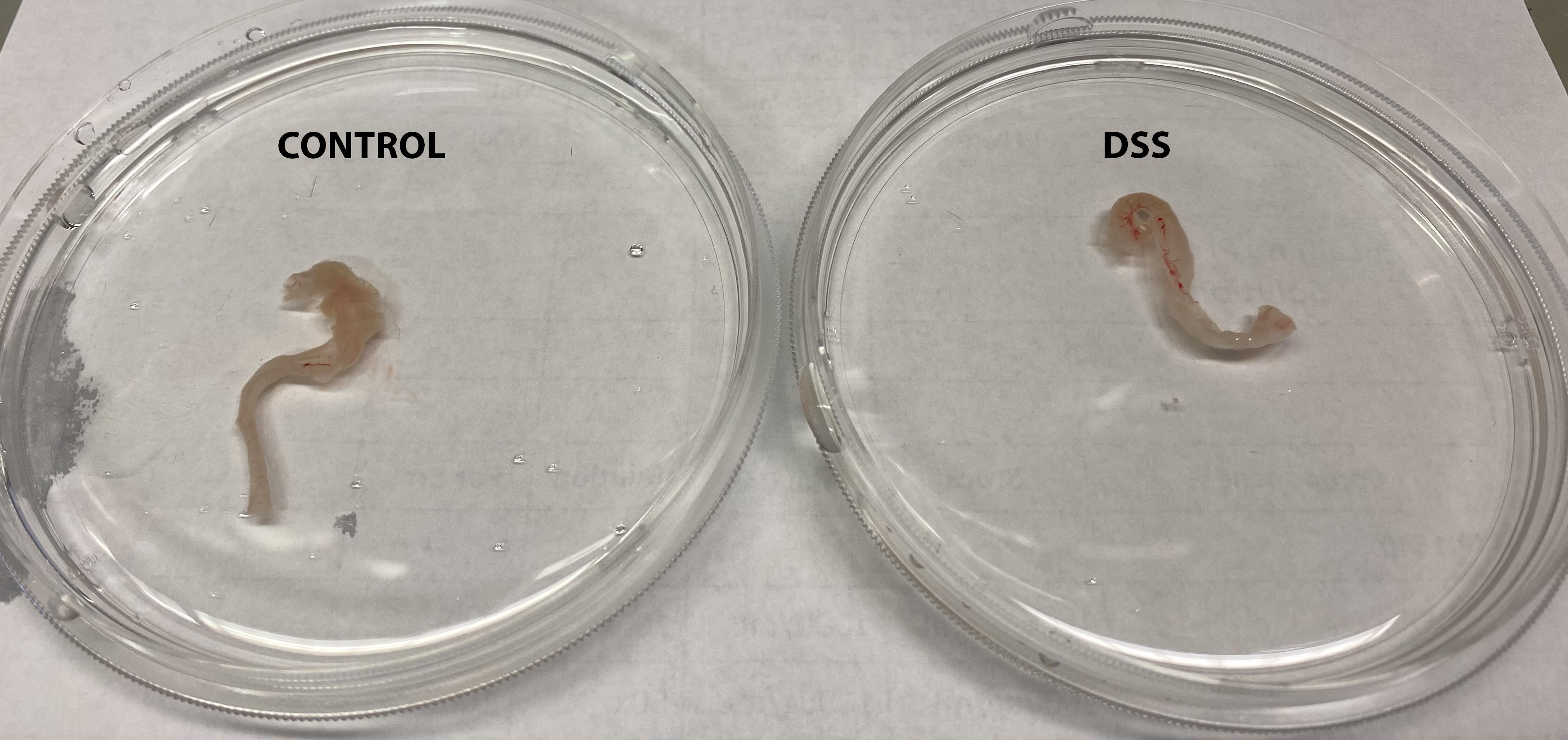
Figure 1. Colon tissues isolated from control or DSS- treated micePlace the colon tissue in a Petri dish containing ice-cold HBSS.
Clean the colon tissue by flushing it with HBSS using a 10 mL syringe with a gavage needle.
Cut the tissue and open it longitudinally. Wash it in the same Petri dish several times by replacing it with fresh ice-cold HBSS.
Once clean, cut the tissue into small pieces (approximately 2 cm long) and keep them with HBSS in the same Petri dish on ice until proceeding to the next step.
Note: Clean and cut all colon tissues required for the experiment before proceeding to the next step.
Isolation of crypts and lamina propria
In this step, the epithelial layer containing the crypts will be isolated from the rest of the tissue.
Transfer the colon pieces into a 50 mL conical tube containing 15 mL of epithelial cell solution.
Incubate at 37 °C in a water bath for 15 min and manually shake the tubes gently every 5 min.
After incubation, transfer the tubes to ice for 10 min.
Transfer the tissue to a new 50 mL conical tube containing 15 mL of fresh HBSS.
Note: It is important to proceed with the crypt isolation (steps B4–B7) using fresh HBSS without EDTA.
Manually shake vigorously for 30–40 s.
Take a small sample of the supernatant, transfer it into a glass slide, and look under the microscope to verify the presence of colon crypts (see example in Figure 2). If crypts are present, proceed to step B7.
Note: If crypts are not present, extend the incubation time in EDTA for 10 more minutes on ice and shake again for 30–40 s.
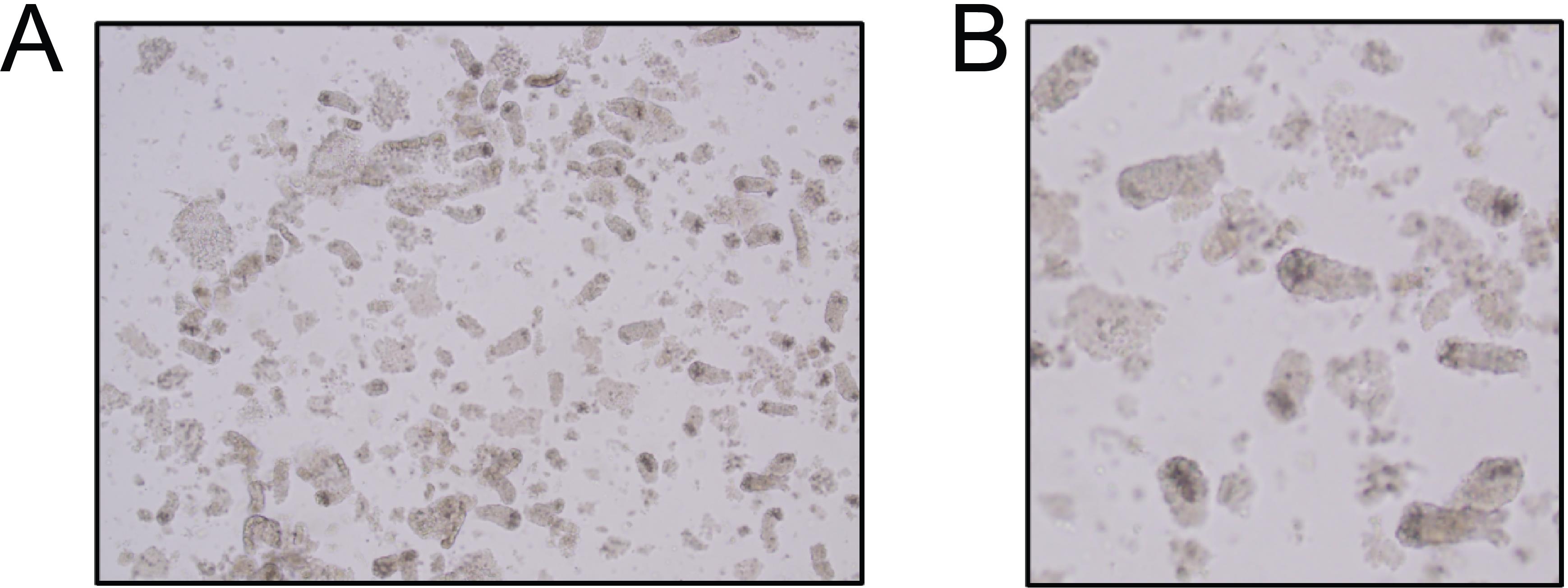
Figure 2. Brightfield image of colon crypts. (A) Colon crypts obtained after EDTA incubation and three consecutive rounds of shaking. (B) Magnification from (A). Squares highlight colon crypts.Collect the supernatant by filtering it through a 100 μm strainer into a new 50 mL conical tube and place the tissue back into the same 50 mL conical tube.
Repeat steps B4–B7 two more times to pool three fractions (total volume of 45 mL).
Centrifuge the supernatant containing the three crypt fractions at 400× g for 5 min.
Resuspend the crypt pellet with 20 mL of epithelial wash buffer and keep the samples on ice until the epithelial fraction digestion step.
Keep the remaining tissue (lamina propria) in a 50 mL conical tube containing 20 mL of HBSS. This tissue will be digested in the next step.
Digestion
Lamina propria fraction
After epithelial layer separation, take the rest of the tissue and place it in the 50 mL conical tube containing 20 mL of HBSS.
Wash the tissue by gently shaking it for 30 s. Discard the HBSS and add 20 mL of new HBSS. Repeat this step at least three times.
Note: This step is very important, as any remaining EDTA will block the lamina propria digestion enzyme.
After washing, cut the tissue into small pieces (approximately 0.5 cm long) and transfer them into a new 15 mL conical tube containing 10 mL of lamina propria solution.
Incubate at 37 °C in a water bath for 20 min.
After 20 min, start pipetting up and down with the 5 mL pipette until the tissue passes smoothly through the pipette.
Note: You can start with the 10 mL pipette and then move to the 5 mL.
Incubate at 37 °C for 10 min.
Start pipetting up and down with a 1 mL pipette until the tissue gets completely dissociated.
Transfer samples to ice and add 1 mL of FBS and 80 μL of EDTA to stop the digestion reaction.
Pipette up and down with a 1 mL pipette and filter through a 40 μm strainer into a 50 mL conical tube.
Add HBSS until 30 mL of volume.
Spin down at 400× g for 10 min.
Remove the supernatant and resuspend the cell pellet with 2 mL of RPMI.
Count cells and measure viability using trypan blue and a cell counter.
Epithelial fraction
Spin down the epithelial fraction (containing crypts) at 400× g for 5 min.
Inspect the cell pellet after the spin down.
Note: It is very important to adjust the volume of TrypLE enzyme used depending on the crypt pellet. As a reference, a pellet similar to the one shown in Figure 3A will be resuspended in 5 (left) or 3 mL (right) of TrypLE.

Figure 3. Examples of cell pellets during the epithelial fraction dissociation. (A) Crypt pellet obtained from a control (left) or dextran sodium sulfate (DSS)-treated (right) mouse in 50 mL tubes. (B) Example of a floating cell pellet after digestion with TrypLE in a 15 mL tube.Resuspend the crypt pellet in 5 mL of pre-warmed TrypLE containing DNase I (100 μg/mL).
Pipette gently up and down until the crypt pellet is dissolved and incubate samples for 5 min at room temperature (RT).
After the 5 min, start pipetting again with a 1 mL pipette for a total period of 10 min.
Note: When there are several samples, keep switching from one to the other every 2–3 min.
Inspect the cells under the microscope to see if they are single cells. If there are still clumps of 5–10 cells, keep pipetting for 5 more minutes.
Note: We do not recommend incubating the epithelial fraction with TrypLE for more than 30 min.
Add 1 mL of FBS into the 5 mL of TrypLE to stop the reaction.
Filter through a 40 μm strainer in a 50 mL conical tube. Add epithelial wash buffer up to 50 mL.
Spin down cells at 400× g for 10 min at 4 °C. Critical point: If the cell pellet is not visible or is floating (see example Figure 3B), after this step we recommend increasing the time and the speed of the centrifugation. We have seen that 10 min at 800× g does not compromise cell viability and helps to pellet the cells.
Discard the supernatant by pipetting. Caution: The pellet can easily detach from the bottom of the tube. It is very important to remove the supernatant carefully. If required, leave 1 or 2 mL of volume to avoid losing cells.
Resuspend the cell pellet in 2 mL of epithelial wash buffer.
Count cells and measure viability using trypan blue and a cell counter.
Validation of protocol
Cell viability
Here, we provide the cell viability and cell count of epithelial and stromal cells obtained at the end of the protocol.
Cell count and viability were obtained using trypan blue and an automated cell counter and listed in Table 1.
Table 1. Cell count and viability at end of protocol.
| Mouse | Cell count (total) | Viability | ||
| Epithelium | Lamina propria | Epithelium | Lamina propria | |
| Mouse 1_Untreated | 520 K | 14.1 M | 83% | 98% |
| Mouse 2_Untreated | 540 K | 12.2 M | 84% | 95% |
| Mouse 3_DSS | 396 K | 17.8 M | 93% | 98% |
| Mouse 4_DSS | 340 K | 15 M | 93% | 96% |
Flow cytometry
By flow cytometry, we characterized different cell types within the epithelial and lamina propria fractions harvested from the colon of naive and DSS-treated mice. As seen in Figure 4A and 4C (top), from viable cells, over 80% of cells are epithelial cells (EPCAM+) in the epithelial fraction from naïve mice (left panel), while in DSS-treated mice (right panel), higher CD45+ immune cells are observed along with a decrease in EPCAM+ cells reflective of active inflammation. We also studied the potential presence of endothelial cells and fibroblasts within the epithelial fraction by gating on CD31 and Podoplanin (PDPN), respectively, in stromal cells identified as CD45 and EPCAM double negative. We noticed that in the epithelial fraction, there is no contamination by CD31+ or PDPN+ cells. In the lamina propria fraction (Figure 4B and 4C, bottom), we observed that most cells are stroma (mainly fibroblasts and, to a lesser extent, endothelial cells) in both naïve and DSS mice with higher immune cell infiltration in the last group.

Figure 4. Flow cytometry gating strategy in the colon. (A and B) Representative flow plot and (C) quantification of cellular subsets (pre-gated on live cells) in the epithelial (A) and lamina propria fractions (B) of naive (left panel) compared to dextran sodium sulfate (DSS)-treated mice (right panel). Data are shown as mean ± SD for three mice in each group. CD45+ = immune cells, EPCAM+ = epithelial cells, CD31+ = endothelial cells and PDPN+ = fibroblast.
Protocol for processing colon biopsies from patients with Crohn’s disease
Materials and reagents
Biological materials
This protocol works equally well for both fresh and cryopreserved biopsies, but for consistency and process efficiency, we routinely use cryopreserved tissue (see General notes). We routinely process two biopsies per subject, to have sufficient cell counts for both sequencing and colonoid line establishment. Biopsies were collected from pediatric patients aged 0–17 years (Table 2) undergoing endoscopy at the Children’s Hospital of Philadelphia between January and December 2021. Prior to sample collection, informed consent was obtained in accordance with Institutional Review Board guidelines (protocol 16-013042). Research biopsies were collected by a trained gastroenterologist from the ascending colon in addition to those for clinical care. Patients with a history of bone marrow transplantation or prior diagnosis of any hemorrhagic, coagulopathic, or aggregation disorders were excluded from this study. Expert pathologists confirmed the diagnoses. Control subjects were evaluated for gastrointestinal complaints but were found to have normal endoscopic and histologic findings.
Table 2. Biopsy donor data
| ID | Age | Sex | Race | Ethnicity |
| CTR-1 | 9 | Male | White | Not Hispanic/Latino |
| CTR-2 | 8 | Male | White | Not Hispanic/Latino |
| CTR-3 | 8 | Female | White | Not Hispanic/Latina |
| Crohn’s-1 | 7 | Female | Asian | Not Hispanic/Latina |
| Crohn’s-2 | 9 | Male | White | Not Hispanic/Latino |
| Crohn’s-3 | 8 | Female | White | Not Hispanic/Latina |
Reagents
CryoStor (CS-10, STEMCELL Technologies, catalog number: 07930)
DPBS (Gibco, catalog number: 14190-136)
Sorbitol (Fisher Bioreagents, catalog number: BP439-500)
Sucrose (Sigma, catalog number: S8501)
BSA (Fisher Bioreagents, catalog number: BP1600-100)
Gentamicin (Gibco, catalog number: 15750-060)
DMEM, high glucose (Corning, catalog number: 10-013-CV)
Advanced DMEM/F12 (Thermo Fisher Scientific, catalog number: 12634028)
FBS (Peak Serum, catalog number: PS-FB1)
EDTA (Quality Biological, catalog number: 351-027-721)
HBSS (Gibco, catalog number: 14175-079)
Antibiotic-Antimycotic (Gibco, catalog number: 15240-062)
HEPES (Gibco, catalog number: 15630-080)
GlutaMAX Supplement (Gibco, catalog number: 35050-061)
Liberase TH (Sigma, catalog number: 5401135001)
DNase I (Roche, catalog number: 10104159001)
IntestiCult organoid growth medium, human (STEMCELL Technologies, catalog number: 06010)
TrypLE Express (Gibco, catalog number: 12605-010)
Trypan Blue solution (Gibco, catalog number: 15250061)
Dead cell removal kit (Miltenyi Biotech, catalog number: 130-090-101) and necessary accessories (see Laboratory supplies)
10× Chromium Next GEM Single Cell 3’ v3.1
Solutions
Liberase TH stock is 13 Wunsch Units (WU)/mL (2.5 mg collagenase/mL) in HBSS. Aliquots are stored at -20 °C, and freeze/thaw is limited to two cycles
DNase I stock is 50 U/mL (25 mg/mL) in DPBS. Aliquots are stored at -20 °C, and freeze/thaw is limited to five cycles
HBSS-BSA (1% BSA, sterilized by filtration)
Biopsy collection medium (see Recipes)
Chelation buffer; prepare fresh every time (see Recipes)
Transport medium (see Recipes)
Thaw medium (see Recipes)
Stroma dissociation enzyme (see Recipes)
Epithelium dissociation enzyme (see Recipes)
Recipes
Biopsy collection medium
Reagent Final concentration Quantity Advanced DMEM/F12 1× 500 mL Antibiotic-antimycotic 1× 5 mL HEPES 10 mM 5 mL GlutaMAX 1× 5 mL Total n/a 515 mL Chelation buffer (sterilize by filtration and chill on ice)
Reagent Final concentration Quantity Sorbitol 2% 0.5 g Sucrose 1% 0.25 g BSA 1% 0.25 g Gentamicin 1% 25 μL DPBS 1× 25 mL Total n/a 25 mL Transport medium
Reagent Final concentration Quantity DMEM 1× 500 mL Antibiotic-Antimycotic 1× 5 mL HEPES 10 mM 5 mL Total n/a 510 mL Thaw medium
Reagent Final concentration Quantity Transport medium n/a 47.5 mL FBS 5% 2.5 mL Total n/a 50 mL Stroma dissociation enzyme (on ice)
Reagent Final concentration Quantity HBSS n/a 5 mL Liberase-TH 0.13 WU/mL 50 μL DNase I 0.5 U/mL 50 μL Total n/a 5 mL Epithelium dissociation enzyme
Reagent Final concentration Quantity TrypLE 1× 1.5 mL DNase I 0.5 U/mL 15 μL Total n/a 1.5 mL
Laboratory supplies
Cryopreservation container (CoolCell, Corning, catalog number: CLS432001 or Mr. Frosty, Fisher Scientific, catalog number: 15-350-50)
Dumont SS forceps, two forceps are needed per specimen (Fine Science Tools, catalog number: 11203-25)
15 and 50 mL conical tubes
35 mm round TC dish (alternatively, a well in a 6-well plate can be used)
40 μm cell strainers
FACS tubes
MACS accessories:
MS Columns (Miltenyi Biotec, catalog number: 130-042-201)
MiniMACS Separator (Miltenyi Biotec, catalog number: 130-042-102) or OctoMACS Separator (Miltenyi Biotec, catalog number: 130-042-109)
Equipment
Tube revolver rotator (Thermo Scientific, catalog number: 88881001)
ThermoMixer (Eppendorf, catalog number: 05-412-501)
Vortex mixer (e.g., Vortex Genie-2, Scientific Industries, catalog number: SI-0236)
Stereomicroscope (Nikon, model: SMZ-U)
Centrifuge supporting 15 and 50 mL conicals
Tabletop microcentrifuge
Hemocytometer for cell number and viability counting
Automated cell counting device (e.g., Invitrogen Countess II)
Procedure
Cryopreservation of biopsies
Two colonic biopsies are collected during colonoscopy into Eppendorf tubes containing 1 mL of biopsy collection medium and delivered to the lab immediately for cryopreservation.
Washing of the tissue fragments (do not use vacuum aspirator):
Note: This is particularly important to prevent contamination if establishing primary culture from the specimen (e.g., patient-derived colonoids).
Remove the biopsy collection medium.
Repeat the following three times: add 1 mL of DPBS, pipette up and down 5–10 times, aspirate the DPBS with a P1000 pipette.
Using a P1000 tip, gently transfer both fragments of tissue into a labeled cryovial containing 1 mL of CryoStor (try to carry over as little PBS as possible) and let the tissue settle to the bottom of the cryovial.
Place the cryovials into the cryopreservation jar.
Place the cryopreservation jar in a -80 °C freezer.
After an overnight incubation at -80 °C, samples should be moved to liquid nitrogen storage.
Thaw the cryopreserved tissue
Thaw the biopsy in a water bath and transfer it to a 1.5 mL Eppendorf tube.
Note: If using a bead bath, place a beaker with pre-heated water into the beads and place the cryovial in the water.
Wash three times with 1 mL of thaw medium.
Here and below, follow this procedure for one wash cycle: add 1 mL of appropriate wash solution (thaw medium here) and pipette up and down 10 times to wash the tissue without letting the fragment enter the pipette tip. Remove the spent wash solution.
Isolation of crypts and lamina propria
Wash twice with 1 mL of chelation buffer (Figure 5B) and discard the spent buffer.
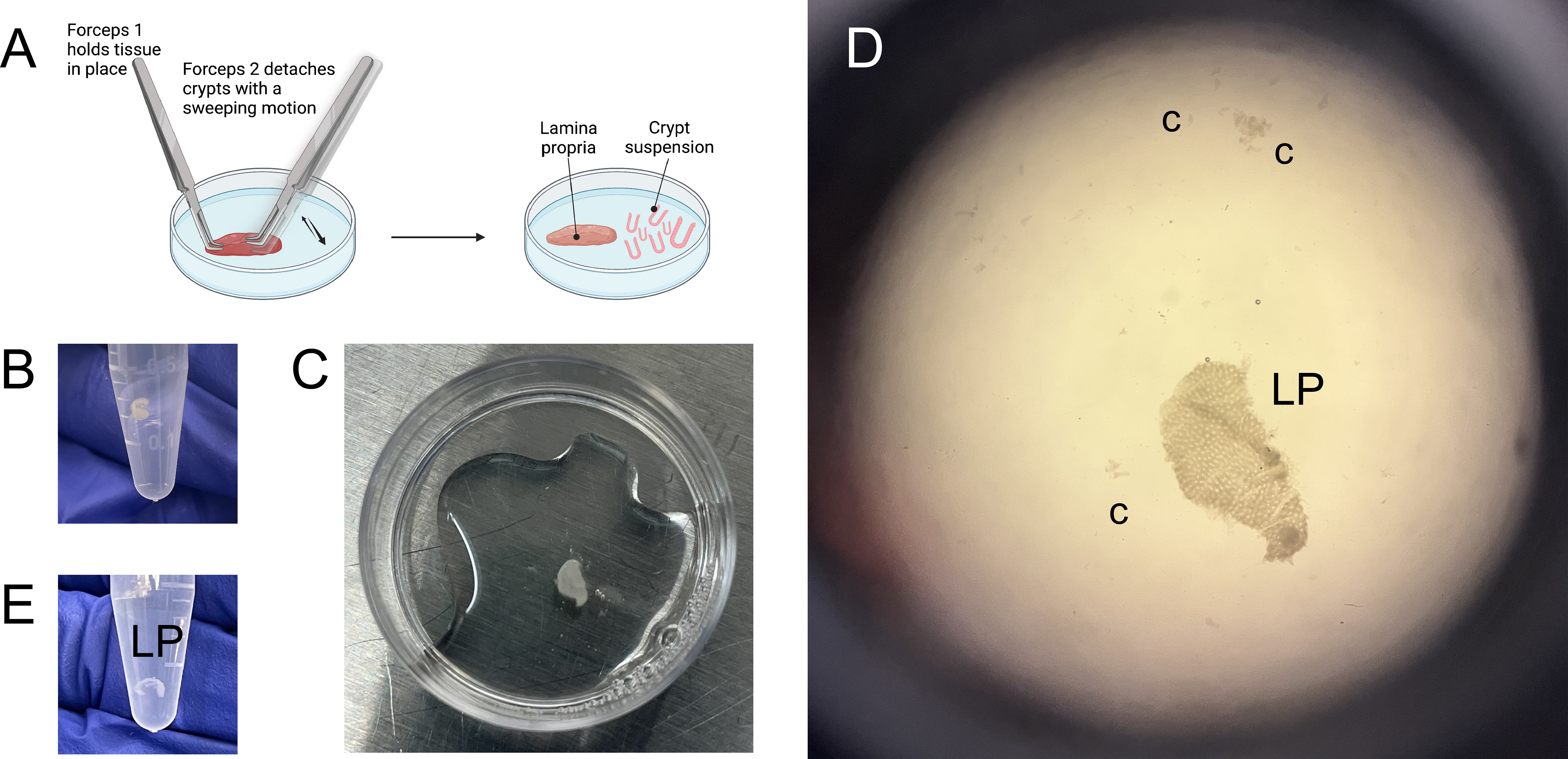
Figure 5. Separation of colonic crypts from lamina propria using forceps. (A) Graphical representation of the crypt scraping process, also remonstrated in Video 1. (B) Colonic biopsy, recovered from cryopreservation and suspended in chelation buffer. (C) Colonic biopsy, after the rotation in chelation buffer and EDTA, post-vortexing and transferred into a 35 mm dish for scraping. (D) View of separated lamina propria (LP) and colonic crypts (c) post-scraping. (E) Lamina propria suspended in DPBS prior to enzymatic digestion.Video 1. Demonstration of crypt scrapingAfter this, add 80 μL of 0.5 M EDTA to the remaining ~20 mL of chelation buffer.
Add 1 mL of chelation buffer + EDTA (final concentration 2 mM) to the tissue in a 1.5 mL Eppendorf tube and rotate cold (in a tube revolver placed in a cold room, set to 10 rpm) for 15 min. During this time, label and chill on ice:
35 mm round TC dish for scraping;
15 mL conical for crypt collection;
1.5 mL Eppendorf tube for stroma collection.
Vortex the tissue on maximum speed setting for five cycles: vortex for 30 s followed by 30 s rest on ice.
Note: For some samples, crypts begin separating during vortexing.
Transfer the spent chelation buffer (may contain crypts) into the prepared 15 mL crypt collection tube.
Use 1 mL of fresh chelation buffer to gently transfer the tissue into a prepared 35 mm dish (Figure 5C).
Under a stereomicroscope, position the biopsy fragment with the luminal side (epithelium) facing up and scrape off the crypts (Figure 5A and 5D; Video 1).
Note: If it is hard to see which side is up, the easiest way is to begin scraping. If no crypts separate, that indicates the lumen is facing down; flip the fragment over and try again.
Transfer the crypt suspension from the 35 mm dish into the 15 mL crypt collection tube.
Move the remaining tissue into the original Eppendorf tube and mark it with stroma (S).
Use 1 mL of thaw medium to rinse the remaining crypts off the stroma fragments and transfer the suspension to the 15 mL crypt collection tube (leave as little liquid with the stroma as possible).
Submerge the stroma in 1 mL of DPBS and place it on ice (Figure 5E).
Use 2 mL of thaw medium to rinse the remaining crypts of the 30 mm dish and transfer the suspension to the 15 mL crypt collection tube.
Spin the crypt collection tube at 700× g for 1 min.
Note: Here and below, it is beneficial, but not critical, to keep the centrifuges chilled to 4 °C.
Transfer the pellet to a 1.5 mL Eppendorf tube and wash with extra HBSS (~500 μL).
Reconstitute the crypt in 1 mL of cold IntestiCult (with 0.5 U/mL DNase).
Optional: count the crypts (typical yield is 200–500 crypts per biopsy fragment).
Place the crypt suspension to gently rotate in the cold room while processing the lamina propria.
Digestion
Lamina propria fraction
Prepare the enzyme mix that will be used for three cycles of digestion.
Cycle 1: Add 500 μL of enzyme mix to stroma fragments.
Transfer the tube to ThermoMixer for 10 min (37 °C, 800 rpm).
During this time, prime a 40 μm strainer, placed in a 50 mL conical, with 2 mL of FBS; this will be the collection tube.
Force the supernatant through the strainer into the collection tube.
Cycle 2: Add 500 μL of fresh enzyme mix to the remaining tissue fragments.
Slowly pipette the tissue up and down 5–10 times through a P1000 tip to enhance breakdown.
Transfer the tube to ThermoMixer for 10 min (37 °C, 800 rpm).
Force the supernatant through the strainer into the same collection tube.
Cycle 3: Add 500 μL of fresh enzyme mix to stroma fragments.
Slowly pipette the tissue up and down 5–10 times through a P1000 tip to enhance breakdown.
Transfer the tube to ThermoMixer for 10 min (37 °C, 800 rpm).
Force the suspension through the strainer.
Use a plunger from a 1 mL insulin syringe to force the remaining fragments through the strainer.
Wash the strainer with 4 mL of thaw medium.
Transfer the contents from the collection tube to a new 15 mL conical.
Note: This step is optional, but the cell pellet is easier to see after the spin when in a 15 mL conical.
Spin at 700× g for 3 min.
Prime the FACS tube by forcing 50 μL of FBS through the strainer cap.
Reconstitute the pellet in 0.5 mL of thaw Medium and strain into the primed FACS tube.
Count the cells and viability using a hemocytometer and trypan blue exclusion, either manually or in an automated device.
Epithelial fraction
Spin down the crypt suspension at 700× g for 1 min.
Reconstitute the crypt pellet in 1.5 mL of TrypLE + 15 μL of DNase I.
For efficient mixing, split into 3 × 1.5 mL Eppendorf tubes each containing 500 μL of the crypt+enzyme suspension.
Transfer the tube to ThermoMixer for 30 min (37 °C, 800 rpm).
Prime the FACS tube by forcing 1 mL of FBS through the strainer cap. Critical: large clumps could be formed at the end of digestion.
Strain into the primed FACS tube and wash the strainer with 1 mL of thaw medium.
Spin at 700× g for 3 min and reconstitute in 500 μL of HBSS-BSA supplemented with 5 μL of DNase I.
Count the cells and viability using a hemocytometer and trypan blue exclusion, either manually or in an automated device.
For single-cell RNA sequencing, preparation of single-cell cDNA libraries and next-generation sequencing are carried out as outlined in the 10× Chromium Next GEM Single Cell 3’ v3.1 user manual.
Data analysis
We aligned and quantified the reads using 10× Cellranger 3.1.0 and built a gene expression matrix by combining all the available samples. Low-quality cells expressing less than 1,000 genes and mitochondrial UMI (Unique Molecular Identifier) greater than 30% of the cell total were discarded. Only the genes that are expressed in more than 100 cells were used for analysis. The UMI counts for each gene were normalized by the cell totals followed by log transformation after addition of a pseudonumber (Bolstad, 2023) to avoid negative and undefined log values. Single-cell RNA sequencing data metrics and viability counts are listed in Table 3 and Table 4, respectively. Using the top 2,000 genes with highest expression variation across the cells, we performed Principal Component Analysis (PCA) on the log-normalized data with R preprocessCore package (Bolstad, 2023). Top 10 dimensions of the PCA explaining the greatest variation in the data were used as input for non-linear dimensionality reduction with Uniform Manifold Approximation and Projection (UMAP) (Becht et al., 2018). We ran clustering on the UMAP coordinates using density clustering function implemented in Monocle (Qiu et al., 2017). Finally, we assigned the cell types for clusters based on the expression patterns of the known cell markers curated from the literature (Figure 6).
Table 3. Single cell data metrics
| ID | Total cells sequenced | Cells removed due to: | Cells post-filtering | ||
| Doublets* | < 500 genes* | > 30% mitochondrial UMI* | |||
| CTR-1 | 6,998 | 75 | 1,712 | 4,318 | 1,977 |
| CTR-2 | 11,880 | 212 | 1,306 | 4,393 | 5,161 |
| CTR-3 | 9,035 | 38 | 5,536 | 7,833 | 1,044 |
| Crohn’s-1 | 6,905 | 61 | 1,524 | 4,514 | 1,658 |
| Crohn’s-2 | 3,997 | 17 | 2,518 | 3,382 | 513 |
| Crohn’s-3 | 11,587 | 211 | 2,285 | 4,529 | 5,267 |
*The total number of cells removed is not always equal to the sum of columns 3–5, because many cells fit several criteria (e.g., the same cell may have < 500 genes and mitochondrial UMI > 30%)
Table 4. Cell and viability counts
| Sample | % viability | Total live cells | ||
| Stroma | Epithelium | Stroma | Epithelium | |
| CTR-1 | 90 | 48 | 620,000 | 58,500 |
| CTR-2 | 77 | 76 | 328,500 | 70,000 |
| CTR-3 | 85 | 42 | 267,000 | 123,000 |
| Crohn’s-1 | 66 | 57 | 126,000 | 141,000 |
| Crohn’s-2 | 90 | 57 | 114,500 | 50,000 |
| Crohn’s-3 | 65 | 78 | 264,000 | 45,000 |
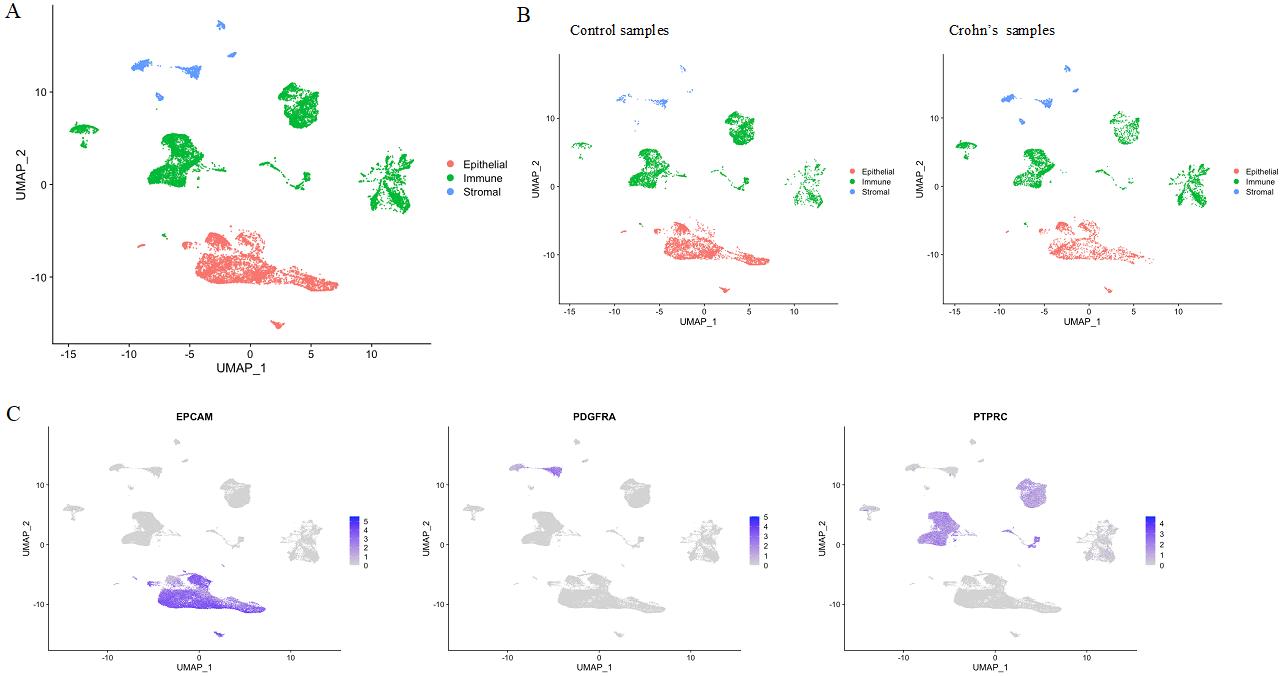
Figure 6. Single-cell RNA-sequencing analysis of tissue biopsies. (A) Uniform Manifold Approximation and Projection (UMAP) visualization of all cells across the six samples color-coded by cell-type compartment. (B) UMAPs divided by disease status. (C) Visualization of the expression of canonical gene markers for epithelial (EPCAM), stromal (PDGFRA), and immune (PTPRC) cells.
Validation of protocol
Cell viability
Single-cell RNA sequencing data metrics and viability counts are listed in Table 3 and Table 4 above.
Note: If the cell viability is lower than 70%, you can use the Dead Cell Removal kit (following the manual provided).
Unsupervised cell clustering
Uniform Manifold Approximation and Projection (UMAP) visualization of all cells across the six samples, as well as visualization of expression of canonical epithelial, stromal, and immune cells are presented above in Figure 6.
General notes and troubleshooting
General notes
The protocols described above are optimized for colon tissues. However, they can also be used for small intestine tissues with some modifications. For murine small intestine, scraping of the villi can be done prior to EDTA incubation to obtain crypt-enriched samples. For human colonic biopsies, the crypt and villi suspension after scraping can be strained through a 100 µm pore membrane to isolate the crypts, while the villi will not go through.
We cryopreserve biopsies prior to processing. Logistically, since we depend on collecting clinical samples from the endoscopy suite, it is difficult to plan the timing and book equipment for fresh tissue processing. Since processing some specimens fresh and some cryopreserved would become a confounding factor, we cryopreserve all specimens and perform the cell isolation protocol in batches.
It is critical that biopsies are cryopreserved as soon as possible, and within 30 min from being procured from a subject.
Mixing back the epithelial cells with stromal cells for single-cell cDNA library generation results in reduced yield of transcriptomes from viable epithelial cells. Therefore, the two fractions must be sequenced separately. To optimize the inevitable cost increase, we sequence biopsies from three subjects at a time: each epithelial fraction results in an independent library, but the stromal fractions can be multiplexed, so there is only one additional library.
Centrifugation steps in the human protocol are short (30 s to 3 min) and thus can be carried out at RT with the sample remaining cool. However, if the centrifuge can be kept at 4 °C, that would further ensure that the sample remains cold.
Troubleshooting
Crypts sticking to forceps: make sure to always dampen the forceps in chelation buffer before touching the tissue.
Poor crypt separation: we find that EDTA stock is the likeliest culprit; opening a new bottle resolves the issue in most cases. Prepare the Chelation buffer fresh every time.
Cell clumps instead of single cells: refresh the enzymes.
Acknowledgments
This publication is part of the Gut Cell Atlas Crohn’s Disease Consortium funded by the Leona M. and Harry B. Helmsley Charitable Trust and is supported by a grant from Helmsley to the Children’s Hospital of Philadelphia. www.helmsleytrust.org/gut-cell-atlas/.
This work is also supported by NIH R01-DK124369 (K.E.H.) and the Children’s Hospital of Philadelphia Gastrointestinal Epithelium Modeling Program.
These protocols were inspired by independent protocols developed by Mahe et al. (2015), Parikh et al. (2019), and Smillie et al. (2019).
Ethical considerations
Animal samples: All animal experiments were performed according to protocols approved by the Institutional Animal Care and Use Committee of the University of Pennsylvania.
Human samples: The institutional review board at the Children’s Hospital of Philadelphia approved the protocol (2014-010826), and all parents of patients provided written informed consent.
References
- Bolstad, B. (2023). preprocessCore: A collection of pre-processing functions. R package version 1.60.2.
- Becht, E., McInnes, L., Healy, J., Dutertre, C. A., Kwok, I. W. H., Ng, L. G., Ginhoux, F. and Newell, E. W. (2018). Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 37(1): 38–44.
- Elmentaite, R., Ross, A. D., Roberts, K., James, K. R., Ortmann, D., Gomes, T., Nayak, K., Tuck, L., Pritchard, S., Bayraktar, O. A., et al. (2020). Single-Cell Sequencing of Developing Human Gut Reveals Transcriptional Links to Childhood Crohn’s Disease. Dev. Cell 55(6): 771–783.e5.
- Mahe, M. M., Sundaram, N., Watson, C. L., Shroyer, N. F. and Helmrath, M. A. (2015). Establishment of human epithelial enteroids and colonoids from whole tissue and biopsy. J. Vis. Exp. (97): 52483.
- Maloy, K. J. and Powrie, F. (2011). Intestinal homeostasis and its breakdown in inflammatory bowel disease. Nature 474(7351): 298–306.
- Ng, S. C., Shi, H. Y., Hamidi, N., Underwood, F. E., Tang, W., Benchimol, E. I., Panaccione, R., Ghosh, S., Wu, J. C. Y., Chan, F. K. L., et al. (2017). Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies. Lancet 390(10114): 2769–2778.
- Parikh, K., Antanaviciute, A., Fawkner-Corbett, D., Jagielowicz, M., Aulicino, A., Lagerholm, C., Davis, S., Kinchen, J., Chen, H. H., Alham, N. K., et al. (2019). Colonic epithelial cell diversity in health and inflammatory bowel disease. Nature 567(7746): 49–55.
- Qiu, X., Mao, Q., Tang, Y., Wang, L., Chawla, R., Pliner, H. A. and Trapnell, C. (2017). Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods 14(10): 979–982.
- Smillie, C. S., Biton, M., Ordovas-Montanes, J., Sullivan, K. M., Burgin, G., Graham, D. B., Herbst, R. H., Rogel, N., Slyper, M., Waldman, J., et al. (2019). Intra- and Inter-cellular Rewiring of the Human Colon during Ulcerative Colitis. Cell 178(3): 714–730.e22.
Article Information
Copyright
© 2023 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Morral, C., Ghinnagow, R., Karakasheva, T., Zhou, Y., Thadi, A., Li, N., Yoshor, B., Soto, G. E., Chen, C. H., Aleynick, D., Weinbrom, S., Fulton, M., Uzun, Y., Bewtra, M., Kelsen, J. R., Lengner, C. J., Tan, K., Minn, A. J. and Hamilton, K. E. (2023). Isolation of Epithelial and Stromal Cells from Colon Tissues in Homeostasis and Under Inflammatory Conditions. Bio-protocol 13(18): e4825. DOI: 10.21769/BioProtoc.4825.
Category
Cell Biology > Single cell analysis > Flow cytometry
Molecular Biology > RNA > RNA sequencing
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








