- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2025
Volume: 15, Issue: 23
Biochemistry
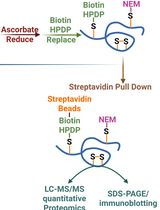
Quantitative Proteomics of Nitrosylated Proteins in Melanoma Using the Biotin-Switch Technique Combined With Tandem Mass Tag Labeling
Bioinformatics and Computational Biology
Detailed Protocol for Segmentation and Quantification of Overlapping Prospore Membranes using DeMemSeg
Biological Sciences
Room-Temperature Storage of Zebrafish and Medaka Sperm Using Lactic Acid-Stabilized L-15 Medium
Biophysics
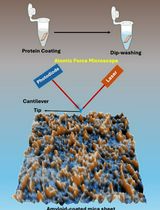
An Optimized Protocol for High-Quality AFM Imaging of Amyloid Fibrils
Immunology
Utilizing EdU to Track Leukocyte Recruitment to the Brain
Microbiology
Imaging the Entire Sexual Life Cycle of the Budding Yeast Saccharomyces cerevisiae Using a Microfluidic Platform
Molecular Biology
Analyzing the Translatome of Lymphatic and Venous Endothelial Cells In Vivo via Translating Ribosome Affinity Purification (TRAP)
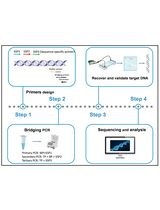
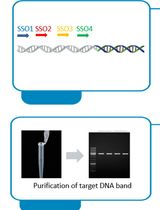
Bridging PCR-Based Genome-Walking Protocol
Implementation of Fusion Primer-Driven Racket PCR Protocol for Genome Walking
Neuroscience
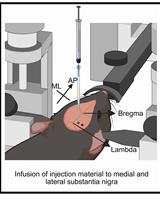
A One-Step Mouse Model of Parkinson’s Disease Combining rAAV-α-Synuclein and Preformed Fibrils of α-Synuclein
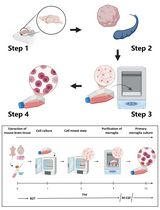
Revisiting Primary Microglia Isolation Protocol: An Improved Method for Microglia Extraction
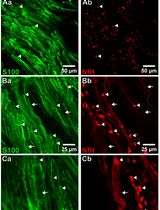
Whole-Mount Immunostaining for the Visual Separation of A- and C-Fibers in the Study of the Sciatic Nerve
Intraepidermal Nerve Fiber Quantification of the Mouse Hind Paw Footpads: A Detailed and Simplified Protocol
Plant Science
Synchronizing Germination Rates Across Plant Species for Fabricated Ecosystems EcoFAB 2.0
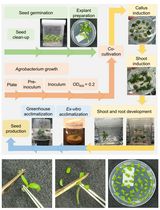
Highly Efficient Agrobacterium-Mediated Transformation of Tomato cv Micro-Tom From Cotyledon Explants
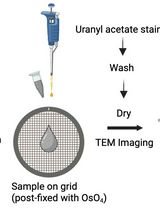
Preparation and Negative Staining for Visualization of Cyanoglobule Lipid Droplets Using Transmission Electron Microscopy
Stem Cell
A Protocol to Induce Brown and Beige Adipocyte Differentiation From Murine and Human Adipose-Derived SVF
A Simplified 3D-Plasma Culture Method for Generating Minimally Manipulated Autologous Equine Muscle-Derived Progenitor Cells