- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2025
Volume: 15, Issue: 5
Biochemistry
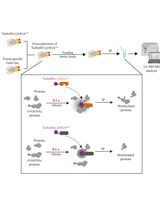
Tissue-Specific Profiling of O-GlcNAcylated Proteins in Drosophila Using TurboID-CpOGAM
Quantification of Total Free Radicals in Drosophila Using a Fluorescence-Based Biochemical Assay
Colorimetric Determination of Tungsten and Molybdenum in Biological Samples
Bioinformatics and Computational Biology
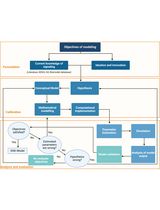
Computational Cellular Mathematical Model Aids Understanding the cGAS-STING in NSCLC Pathogenicity
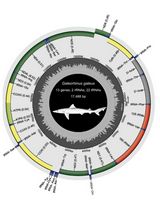
Annotated Bioinformatic Pipelines for Genome Assembly and Annotation of Mitochondrial Genomes
Annotated Bioinformatic Pipelines for Phylogenomic Placement of Mitochondrial Genomes
Developmental Biology
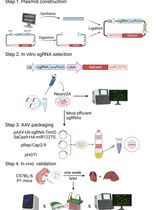
Cardiac-Specific Gene Editing via an AAV9-Tnnt2-SaCas9-miR122TS Vector
Immunology
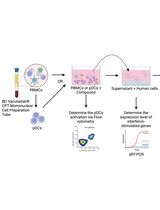
Protocol for Screening Host-Targeting Antivirals (HTAs) Using Human PBMCs and pDCs
Microbiology
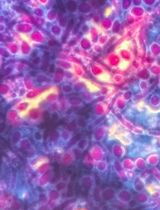
Microbial Biofilm Detection and Differentiation by Dual Staining Using Maneval’s Stain
Integrated Co-extraction Protocol for Transcriptomic and 1H NMR Metabolomic Analysis of Multi-species Biofilms
Neuroscience
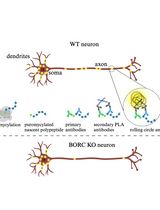
Puromycin Proximity Ligation Assay (Puro-PLA) to Assess Local Translation in Axons From Human Neurons
Monitoring Changes in Intracellular Chloride Levels Using the FRET-Based SuperClomeleon Sensor in Organotypic Hippocampal Slices
Procedure for Reliable and Long-Lasting Ex Vivo Recordings of Sciatic Nerve Activity in Mice
Plant Science
Isolation and Biophysical Characterization of Extracellular Vesicles From Hairy Root Cultures
An Activity-Based Proteomics with Two-Dimensional Polyacrylamide Gel Electrophoresis (2D-PAGE) for Identifying Target Proteases in Arabidopsis Apoplastic Fluid
Stem Cell
Differentiation, Maintenance, and Contraction Profiling of Human Induced Pluripotent Stem Cell–Derived Cardiomyocytes
Systems Biology
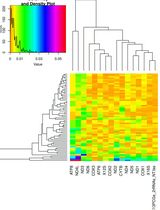
Automated Sequential Derivatization for Gas Chromatography-[Orbitrap] Mass Spectrometry-based Metabolite Profiling of Human Blood-based Samples













![Automated Sequential Derivatization for Gas Chromatography-[Orbitrap] Mass Spectrometry-based Metabolite Profiling of Human Blood-based Samples](https://en-cdn.bio-protocol.org/imageup/arcimg/20250107224024942.jpg?t=1767769488)