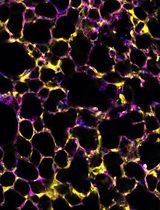
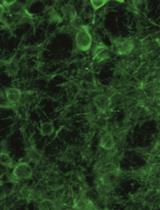
Imaging Single-Cell Ca2+ Dynamics of Brainstem Neurons and Glia in Freely Behaving Mice
In vivo brain imaging, using a combination of genetically encoded Ca2+ indicators and gradient refractive index (GRIN) lens, is a transformative technology that has become an increasingly potent research tool over the last decade. It allows direct visualisation of the dynamic cellular activity of deep brain neurons and glia in conscious animals and avoids the effect of anaesthesia on the network. This technique provides a step change in brain imaging where fibre photometry combines the whole ensemble of cellular activity, and multiphoton microscopy is limited to imaging superficial brain structures either under anaesthesia or in head-restrained conditions. We have refined the intravital imaging technique to image deep brain nuclei in the ventral medulla oblongata, one of the most difficult brain structures to image due to the movement of brainstem structures outside the cranial cavity during free behaviour (head and neck movement), whose targeting requires GRIN lens insertion through the cerebellum—a key structure for balance and movement. Our protocol refines the implantation method of GRIN lenses, giving the best possible approach to image deep extracranial brainstem structures in awake rodents with improved cell rejection/acceptance criteria during analysis. We have recently reported this method for imaging the activity of retrotrapezoid nucleus and raphe neurons to outline their chemosensitive characteristics. This revised method paves the way to image challenging brainstem structures to investigate their role in complex behaviours such as breathing, circulation, sleep, digestion, and swallowing, and could be extended to image and study the role of cerebellum in balance, movement, motor learning, and beyond.Key features• We developed a protocol that allows imaging from brainstem neurons and glia in freely behaving rodents.• Our refined method of GRIN lenses implantation and cell sorting approach gives the highest number of cells with the least postoperative complications.• The revised deep brainstem imaging method paves way to understand complex behaviours such as cardiorespiratory regulation, sleep, swallowing, and digestion.• Our protocol can be implemented to image cerebellar structures to understand their role in key functions such as balance, movement, motor learning, and more.Graphical overview