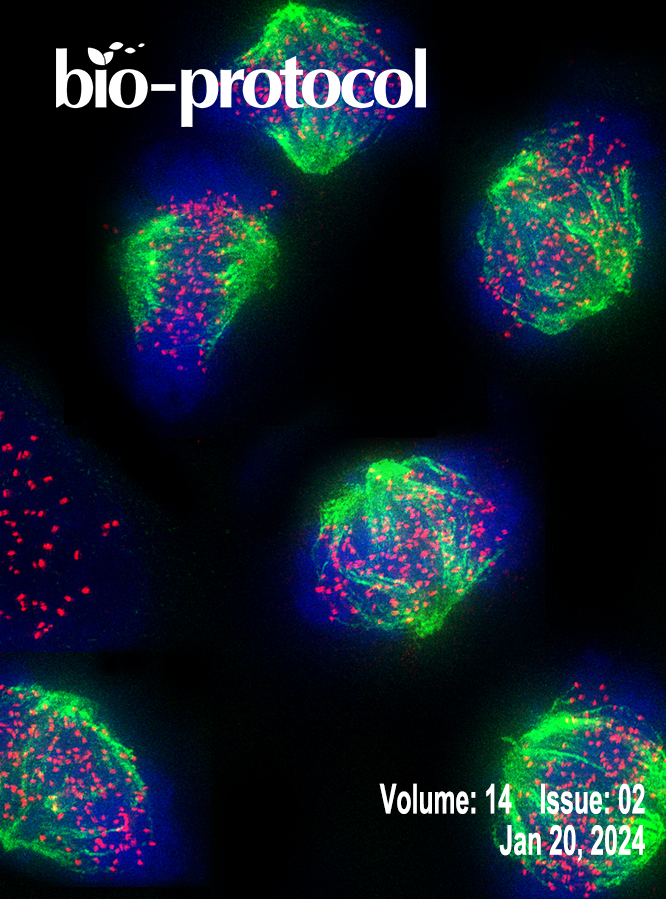
Isolation and Enrichment of Major Primary Neuroglial Cells from Neonatal Mouse Brain
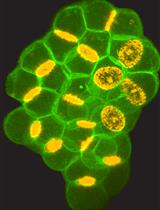
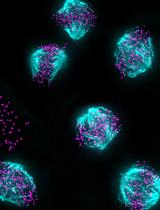
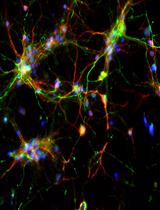
The central nervous system (CNS) relies on the complex interaction of neuroglial cells to carry out vital physiological functions. To comprehensively understand the structural and functional interplay between these neuroglial cells, it is essential to establish an appropriate in vitro system that can be utilized for thorough investigation. Traditional protocols for establishing primary neuronal and mixed glial cultures from prenatal mice or neural stem cells require sacrificing pregnant mice and have the drawback of yielding only specific types of cells. Our current protocol overcomes these drawbacks by utilizing the brain from day-0 pups to isolate CNS resident neuroglial cells including astrocytes, microglia, oligodendrocytes [oligodendrocyte precursor cells (OPCs) and differentiated oligodendrocytes], and meningeal fibroblasts, as well as hippocampal neurons, avoiding sacrificing pregnant mice, which makes this procedure efficient and cost effective. Furthermore, through this protocol, we aim to provide step-by-step instructions for isolating and establishing different primary neuroglial cells and their characterization using cell-specific markers. This study presents an opportunity to isolate, culture, and establish all major CNS resident cells individually. These cells can be utilized in various cell-based and biochemical assays to comprehensively investigate the cell-specific roles and behaviors of brain resident cells in a reductionist approach.Key features• Efficient isolation of major neuroglial cells like meningeal fibroblasts, neurons, astrocytes, oligodendrocytes, and microglia from a single day-0 neonatal mouse pup’s brain.• Circumvents the sacrifice of pregnant female mice.• Acts as a bridging experimental method between secondary cell lines and in vivo systems.• Isolated cells can be used for performing various cell-based and biochemical assays.Graphical overviewSteps for isolation of meningeal fibroblast and neuroglial cells from day 0 pups of mice (Created using BioRender.com)