A Miniature Sucrose Gradient for Polysome Profiling
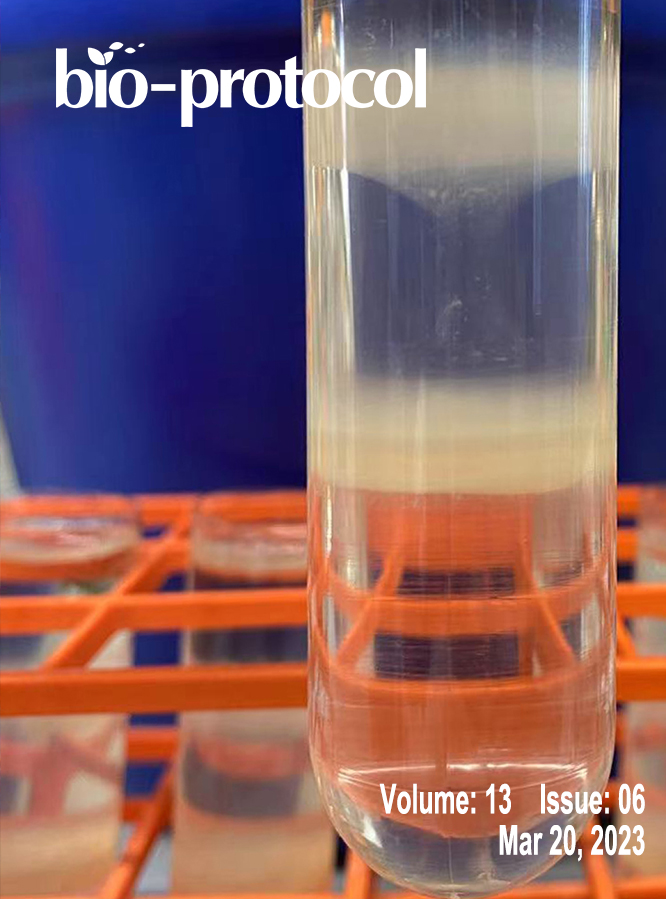
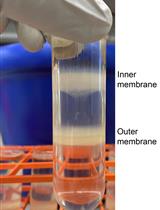
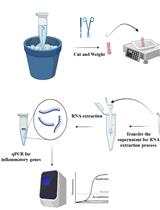
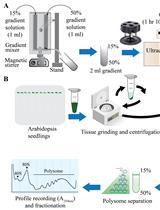
Polysome profiling by sucrose density gradient centrifugation is commonly used to study the overall degree of translation (messenger RNA to protein synthesis). Traditionally, the method begins with synthesis of a 5–10 mL sucrose gradient onto which 0.5–1 mL of cell extract is layered and centrifuged at high speed for 3–4 h in a floor-model ultracentrifuge. After centrifugation, the gradient solution is passed through an absorbance recorder to generate a polysome profile. Ten to twelve fractions (0.8–1 mL each) are collected for isolating different RNA and protein populations. The overall method is tedious and lengthy (6–9 h), requires access to a suitable ultracentrifuge rotor and centrifuge, and requires a substantial amount of tissue material, which can be a limiting factor. Moreover, there is often a dilemma over the quality of RNA and protein populations in the individual fractions due to the extended experiment times. To overcome these challenges, here we describe a miniature sucrose gradient for polysome profiling using Arabidopsis thaliana seedlings that takes ~1 h centrifugation time in a tabletop ultracentrifuge, reduced gradient synthesis time, and also less tissue material. The protocol described here can be easily adapted to a wide variety of organisms and polysome profiling of organelles, such as chloroplasts and mitochondria.Key Features• Mini sucrose gradient for polysome profiling that requires less than half the processing time vs. traditional methods.• Reduced starting tissue material and sample volume for sucrose gradients.• Feasibility of RNA and protein isolation from polysome fractions.• Protocol can be easily modified to a wide variety of organisms (and even polysome profiling of organelles, such as chloroplast and mitochondria).Graphical OverviewFigure 1. Graphical overview of polysome profiling using mini sucrose gradient. A. One milliliter each of 15% (w/v) and 50% (w/v) sucrose gradient solution is added to the individual chambers of the gradient maker. While mixing with a small magnetic stirrer in the 50% solution chamber, base station knob is turned to open position, allowing sucrose gradient solution to slowly flow through the outlet into a 2.2 mL gradient tube. After centrifugation at 50,000 rpm (213,626.2 × g) in a swinging bucket rotor for 70 min at 4 °C, the gradient tube is stored at 4 °C for the next steps. B. Cell extract from 12-day-old vertically grown Arabidopsis thaliana seedlings is centrifuged twice and 100 µL of supernatant is gently layered on the pre-made sucrose gradient from step A. After centrifugation as described in step A, polysome profile is obtained by feeding the gradient solution through an absorbance recorder (A254 nm). Eight (200 µL) fractions are collected for RNA and protein isolation.