- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2022
Volume: 12, Issue: 17
Biochemistry
Split-Chloramphenicol Acetyl Transferase Assay to Study Protein-Protein Interactions and Ubiquitylation in Escherichia coli
Production, Purification, and Fluorometric Activity Assay of Human Aldehyde Dehydrogenases
Biological Engineering
Incorporation of a Chemically Diverse Set of Non-Standard Amino Acids into a Gram-Positive Organism
Cancer Biology
Microscopic Detection of DNA Synthesis in Early Mitosis at Repetitive lacO Sequences in Human Cells
Detection of Alternative End-Joining in HNSC Cell Lines Using DNA Double-Strand Break Reporter Assays
Editorial
Immunology
High-Efficiency Retroviral Transduction for Type 1 Regulatory T Cell Differentiation
Microbiology
A New Tool for the Flexible Genetic Manipulation of Geobacillus kaustophilus
Molecular Biology
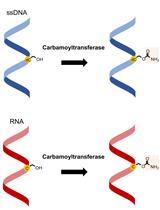
Carbamoyltransferase Enzyme Assay: In vitro Modification of 5-hydroxymethylcytosine (5hmC) to 5-carbamoyloxymethylcytosine (5cmC)
Neuroscience
Evaluation of Mitochondrial Turnover Using Fluorescence Microscopy in Drosophila
Stem Cell
Generation of iMyoblasts from Human Induced Pluripotent Stem Cells