- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Negative Ion Mode nanoLC-ESI-MS/MS Analyses of Permethylated Sulfated Glycans
Published: Vol 10, Iss 10, May 20, 2020 DOI: 10.21769/BioProtoc.3618 Views: 4369
Reviewed by: Manjula MummadisettiAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Permethylation and Microfractionation of Sulfated Glycans for MS Analysis
Shin-Yi Yu [...] Kay-Hooi Khoo
May 20, 2020 4653 Views

Bioorthogonal Labeling and Chemoselective Functionalization of Lung Extracellular Matrix
Zihan Ling [...] Xi Ren
Feb 20, 2021 4386 Views

Identification of Matriglycan by Dual Exoglycosidase Digestion of α-Dystroglycan
Ishita Chandel and Kevin P. Campbell
Sep 20, 2023 1336 Views
Abstract
We have developed enabling techniques for sulfoglycomics based on MALDI-MS mapping and MS/MS sequencing of permethylated sulfated glycans. We then extended further the analytical workflow to C18 reverse phase (RP)-nanoLC-nanoESI-MS/MS analyses of permethylated sulfated glycans in the negative ion mode. The advantages are that extra sulfates on permethylated di- and multiply sulfated glycans will survive in nanoESI conditions to allow detection of multiply charged intact molecular ions, and more comprehensive MS/MS can be performed in an automated fashion at higher sensitivity, compared with MALDI-MS/MS. Parallel higher energy collision dissociation (HCD) and ion trap collision induced dissociation (CID)-based MS2, coupled with product-dependent MS3 in data dependent acquisition mode proved to be highly productive when applied to resolve and identify the isomeric sulfated glycan structures. In-house glycomic data mining software, GlyPick, was developed and used to automate the downstream process of identification and relative quantification of target sulfated glycotopes based on summed intensity of their diagnostic MS2 ions extracted from thousands of HCD-MS2 and/or CID-MS2 data.
Keywords: Permethylated sulfated glycansBackground
Current mass spectrometry (MS)-based glycomic mapping remains insufficient to delineate the full complexity of the glycome. Although MALDI-MS mapping and MS/MS sequencing would efficiently afford a very useful first impression glycomic profile, it is nearly impossible to acquire MALDI-MS/MS on every putative glycan signal detected, especially those of low intensities and/or occurring at higher masses. In that respect, LC-ESI-MS/MS analysis in an automated and data-dependent acquisition (DDA) mode provides a far more comprehensive MS/MS data coverage (Cheng et al., 2013; Cheng et al., 2015; Hsiao et al., 2017; Yu et al., 2018). We have demonstrated that permethylation in conjunction with 2 steps C18/amine SPE or a single step mixed mode MAX SPE fractionation can yield fully methylated non-sulfated, mono-sulfated, and di-sulfated glycans in separate pools (Yu et al., 2020) for MALDI-MS screening. We found that loss of sodium sulfite from permethylated di-sulfated glycans occurred readily during MALDI-ionization (Figure 1A), while the two sulfates on permethylated glycans were fully retained when analyzed by nanoESI-MS in negative ion mode, allowing them to be detected as [M-2H]2- (Figure 1B), and further selected for MS/MS analyses at high sensitivity. Moreover, we showed that the negative mode nanoLC-MS/MS analysis of permethylated sulfated glycans on a C18 reverse phase capillary column could be efficiently carried out using the common acetonitrile/formic acid/water solvent system, and hence be fully compatible with the normal set-up of an analytical laboratory devoted to proteomics.
By virtue of a panel of synthetic sulfated glycans, we have shown that the beam-type higher energy collision dissociation (HCD) MS/MS as implemented on the hybrid Orbitrap series would afford a range of low mass fragment ions. These diagnostic ions would define the location of sulfate on which glycosyl residue at which position (Cheng et al., 2015). Ion trap-based CID MS2, on the other hand, would not retain these very useful ions due to its one third low mass cut-off (Patnode et al., 2013). It does, however, offer a higher sensitivity and acquisition rate, allowing definitive assignment of target sulfated glycotopes via MS2 product ion-dependent MS3. Occasionally, when a di-sulfated O-glycan may carry a different permutation of sulfated glycotopes on either the 6-arm or the extended 3-arm, additional targeted MSn analyses can help identify the existence of isomers (Yu et al., 2018). Moving from the original linear ion trap-Orbitrap hybrid systems to more recent tribrid Orbitrap Fusion systems, the current MS systems afford a higher degree of flexibility for different combination of single to multiple stages of HCD versus ion trap-based CID MS/MS to be acquired either in Orbitrap for greater resolution and mass accuracy, or the ion trap for better sensitivity and speed. These aspects will not be further dealt with here. Suffice to point out that the basic principle remains the same, namely to acquire as many HCD/CID MS2 within an elution time window-compatible DDA duty cycle and to couple each, if possible and desirable, to a pre-determined list of product-dependent MS3 for greater depth of structural details. With or without MS3, it is important that the low mass ions produced in negative mode MS2 be retained and detected at reasonably high resolution and mass accuracy (< 5 ppm, if possible).
Online LC and data-dependent MS/MS acquisition will produce a huge dataset which is near impossible to manually analyze systematically. Unlike proteomics, neither the glycan database nor the MS/MS sequencing algorithm is well developed to allow direct MS2 ions or spectral matching search for unambiguous identification. An in-house computational tool, GlyPick, was developed to filter out bona fide glycan MS2 spectra by user-defined criteria, usually by presence of at least 2 to 3 diagnostic glycan fragment ions. It can also extract out and compute the occurrence and summed intensity of a list of user-input diagnostic MS2 ions that will define the presence and relative abundance of those important glycotopes such as 6-sulfated GlcNAc, 3’-sulfated Gal, 6’-sulfated Gal, sulfated LacNAc, sulfated Le, etc, from these MS2 spectra (Hsiao et al., 2017; Yu et al., 2018). Results are output in CSV format, which can be conveniently interrogated using Excel for further data mining and graph plotting.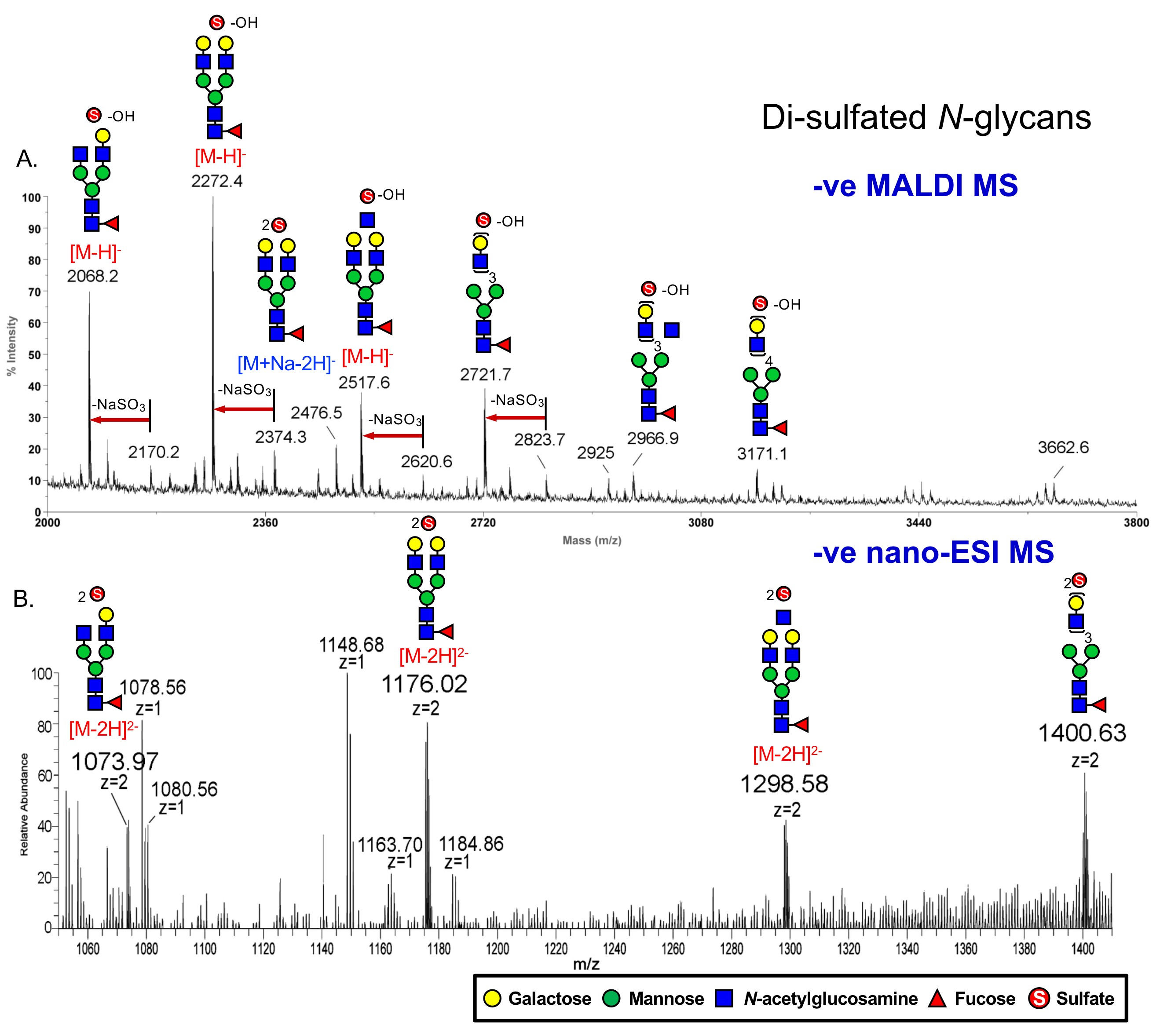
Figure 1. MS spectra of di-sulfated permethylated N-glycans acquired by different ionizations, which are (A) MALDI-MS and (B) nanospray ESI-MS, both in the negative ion mode.
Materials and Reagents
- Plastic pipette tips
- Microcentrifuge tube
- ZipTipC18 (Merck Millipore, catalog number: ZTC18S096 )
- Acetonitrile with 0.1% (v/v) Formic Acid for LC-MS, JT Baker® (VWR, catalog number: JT9832-2 )
- Water with 0.1% (v/v) Formic acid, BAKER ANALYZEDTM, JT Baker® (VWR, catalog number: JT9826-3 )
Equipment
- SpeedVac
- nanoACQUITY UPLC system (Waters Corporation)
- nanoACQUITY M-Class BEH130 C18 column, 1.7 μm, 75 μm x 250 mm (Waters Corporation, catalog number: 186003545 )
- Picoview nanospray source 550 (New Objective, catalog number: PV-550 )
- A hybrid LTQ-Orbitrap EliteTM Mass Spectrometer (Thermo Scientific), or any equivalent high resolution/mass accuracy MS system including the tribrid Orbitrap Fusion (Thermo Fisher Scientific)
Software
- MasslynxTM software v. 4.1 (Waters)
- XcaliburTM software v2.2 (Thermo Fisher Scientific)
- GlyPick (in-house, available upon request)
Procedure
- Sample preparation
- Before subjecting to UPLC system, permethylated glycan sample should be cleaned up further by ZipTipC18 (described in Yu et al., 2020), and dried by SpeedVac.
- Re-dissolve the permethylated glycan sample in 10 μl of 5% acetonitrile, 0.1% formic acid.
- Set up the UPLC system parameters
- Connect the nanoACQUITY UPLC system to an LTQ-Orbitrap EliteTM hybrid mass spectrometer via PicoView nanospray source for nanoLC separation at 35 °C, using a 75 μm ID, 25 cm length C18 BEH column packed with 1.7 μm particles with a 300 Å pore size.
- Set up the constant flow rate of 300 nl/min
- Set the solvent system containing 100% water with 0.1% formic acid (FA) for mobile phase A, and 100% acetonitrile with 0.1% FA for mobile phase B. Use a linear gradient of 30-60% of B over the course of 30 min and then increase to 80% acetonitrile over the course of 5 min and hold isocratically for another 10 min (shown in Figure 2).
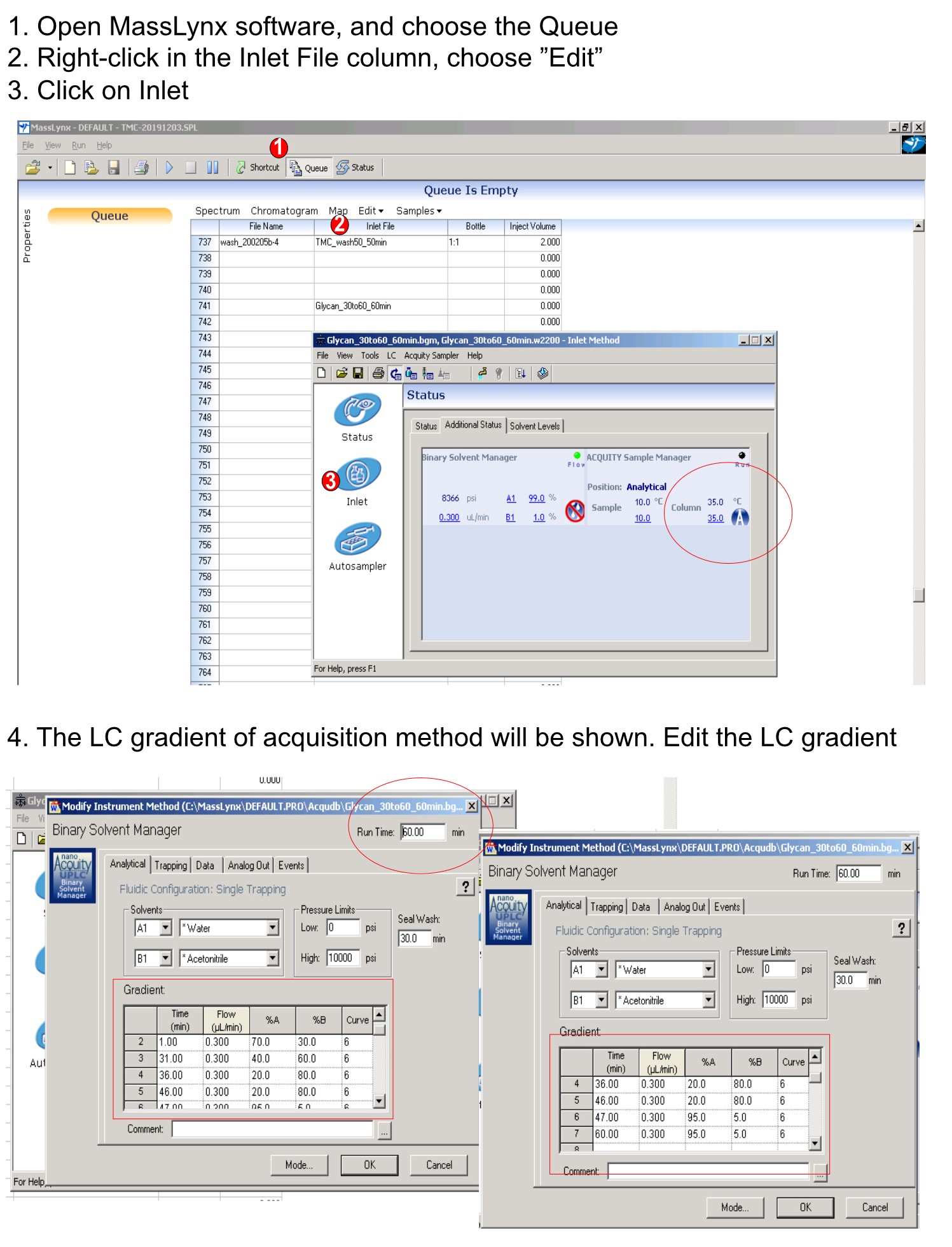
Figure 2. Steps in setting up HPLC gradient method
- Set up parameters for MS acquisition (shown in Figure 3)
- Double-click the Xcalibur software and then, click the Instrument Setup.
- Click “Nth order double play”.
- Choose “Initialize method with Orbitrap Elite support”, and type “5” in Analyze Top N peaks, then click “OK”. The page named “Thermo Xcalibur Instrument Setup” will be shown.
- Set up parameters which are
- Acquire time is set for 60 min and type 3 in the “Scan events” in Segment 1 settings (indicated by blue arrow).
Note: The acquisition time can be adjusted when LC gradient is changed or different UPLC system/reverse phase capillary column is used. - Click Scan Event 1 (active window shown as yellow background).
- Scan Description: Analyzer: FTMS; Mass Range: Normal; Resolution: 120,000; Scan Type: Full; Polarity: Negative; Data type: Profile.
- Scan Ranges: First Mass (m/z) 650; Last Mass (m/z) 2000.
- Click Scan Event 2 (active window shown as yellow background).
- Scan Description: Analyzer: Ion Trap; Mass Range: Normal; Scan type: Normal; Data type: Centroid
- Click Settings and the window “Data Dependent Settings” will be shown.1)Choose “Dynamic Exclusion”; Click “Enabled”; Type Repeat count: 2; Repeat duration (s): 15; Exclusion list size: 500; Exclusion duration (s): 60; Exclusion mass width, choose “By mass”; Low: 1.5; High: 1.5, Click “OK”.2)Choose “Charge State”; Click “Enable charge state screening” and “Enable monoisotopic precursor selection”; Click “Enabled” and choose “Reject charge state”: “2”, “3”, “4 and up”. Click “OK”.
Note: This setting is for mono-sulfated permethylated glycans due to singly charge of mono-sulfated glycans in the negative ion mode.3)Choose “Current Scan Event”; Type Minimum signal threshold (counts): 5000. Click “OK”.4)Choose “Activation”; Choose Activation type: CID; Default charge state: 2; Isolation width (m/z): 2; Normalized collision energy: 35; Activation Q: 0.25; Activation time (ms): 10. Click “OK”.
- Click Scan Event 3 (active window shown as yellow background)
- Scan Description: Analyzer: FTMS; Mass Range: Normal; Resolution: 15,000; Data type: Centroid.
- Click Settings and the window “Data Dependent Settings” will be shown.1)Choose “Dynamic Exclusion”; Click “Enabled”; Repeat count: 2; Repeat duration (s): 15; Exclusion list size: 500; Exclusion duration (s): 60; Exclusion mass width, choose “By mass”; Low: 1.5; High: 1.5. Click “OK”.2)Choose “Charge State”; Click “Enable charge state screening” and “Enable monoisotopic precursor selection”; Click “Enabled” and choose “Reject charge state”: “2”, “3”, “4 and up”. Click “OK”.
Note: This setting is for mono-sulfated permethylated glycans due to singly charge of mono-sulfated glycans in the negative ion mode.3)Choose “Current Scan Event”; Type Minimum signal threshold (counts): 5000. Click “OK”.4)Choose “Activation”; Choose Activation type: HCD; Default charge state: 2; Isolation width (m/z): 2; Normalized collision energy: 110; Activation time (ms): 0.1. Click “OK”.
- Acquire time is set for 60 min and type 3 in the “Scan events” in Segment 1 settings (indicated by blue arrow).
- Go to Tune plus window, and choose Setup→FT Injection Control or click
 in the Instrument Control toolbar. The window named “Injection Control” will be shown. Choose FT, AGC Target Settings, Type “Full MS”: 1.00e6; SIM: 5.00e4; MSn: 5.00e4; Click “Enable Full Scan Injection Waveforms”. Choose Ion Trap, Full MS: 3.00e4; SIM: 1.00e4; MSn: 1.00e4; Zoom: 3000, Click “OK”.
in the Instrument Control toolbar. The window named “Injection Control” will be shown. Choose FT, AGC Target Settings, Type “Full MS”: 1.00e6; SIM: 5.00e4; MSn: 5.00e4; Click “Enable Full Scan Injection Waveforms”. Choose Ion Trap, Full MS: 3.00e4; SIM: 1.00e4; MSn: 1.00e4; Zoom: 3000, Click “OK”.
- Scan ranges in the scan event 1 will be changed to 1,000-4,000 for permethylated mono-sulfated N-glycans due to singly charged property.
- Check the section “Data dependent settings” → “Segment” → “Charge State”, The rejected charge state should be adjusted depends on charge states of glycans. For di-sulfated N- or O-glycans detected in the negative ion mode are normally doubly charged. The rejected charge states should be 1, 3, and 4-and up.
- One should always optimize and calibrate the LC-MS/MS system accordingly.
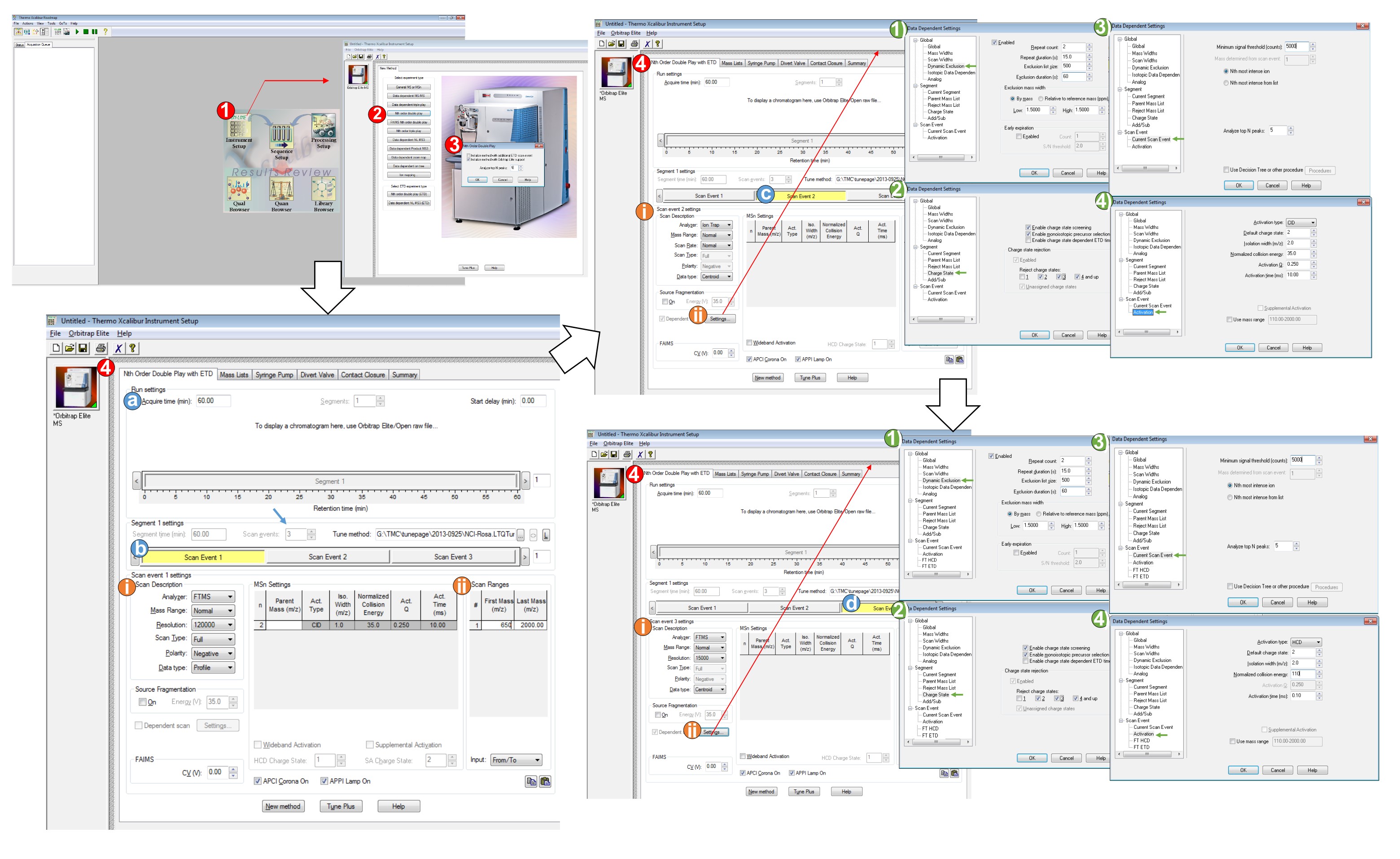
Figure 3. Setting up the MS acquisition
Data analysis
- ESI-MS analysis in the negative ion mode
All data were processed by Xcalibur software v2.2 manually. We observed that permethylated mono-sulfated glycans were eluted earlier than di-sulfated glycans by using the nanoACQUITY UPLC BEH130 column and acetonitrile/formic acid/ water solvent system. The ion signals of mono-sulfated N- and O-glycans were detected as [M-H]-, whereas di-sulfated N- and O-glycans were detected as [M-2H]2- in the ESI-MS spectra. - ESI-MS/MS analyses in the negative ion mode
The acquired HCD-MS2 and CID-MS2 spectra are typically averaged over a period of retention time according to the elution profile of its precursor, and then interpreted manually. In general, fragmentation pattern of permethylated sulfated glycans in the negative ion mode is similar to fragmentation pattern in the positive ion mode (Yu et al., 2006; Hsiao et al., 2017), except that only fragment ions retaining the sulfate and hence the negative charge will be detected in the negative ion mode. Considerable expertise and experience are required to correctly and fully assign the various cleavage ions, while identification of the few well-established diagnostic ions is more straightforward (summarized in Table 1). An example is shown below to illustrate the characteristic fragmentation patterns that can be expected (Figure 4).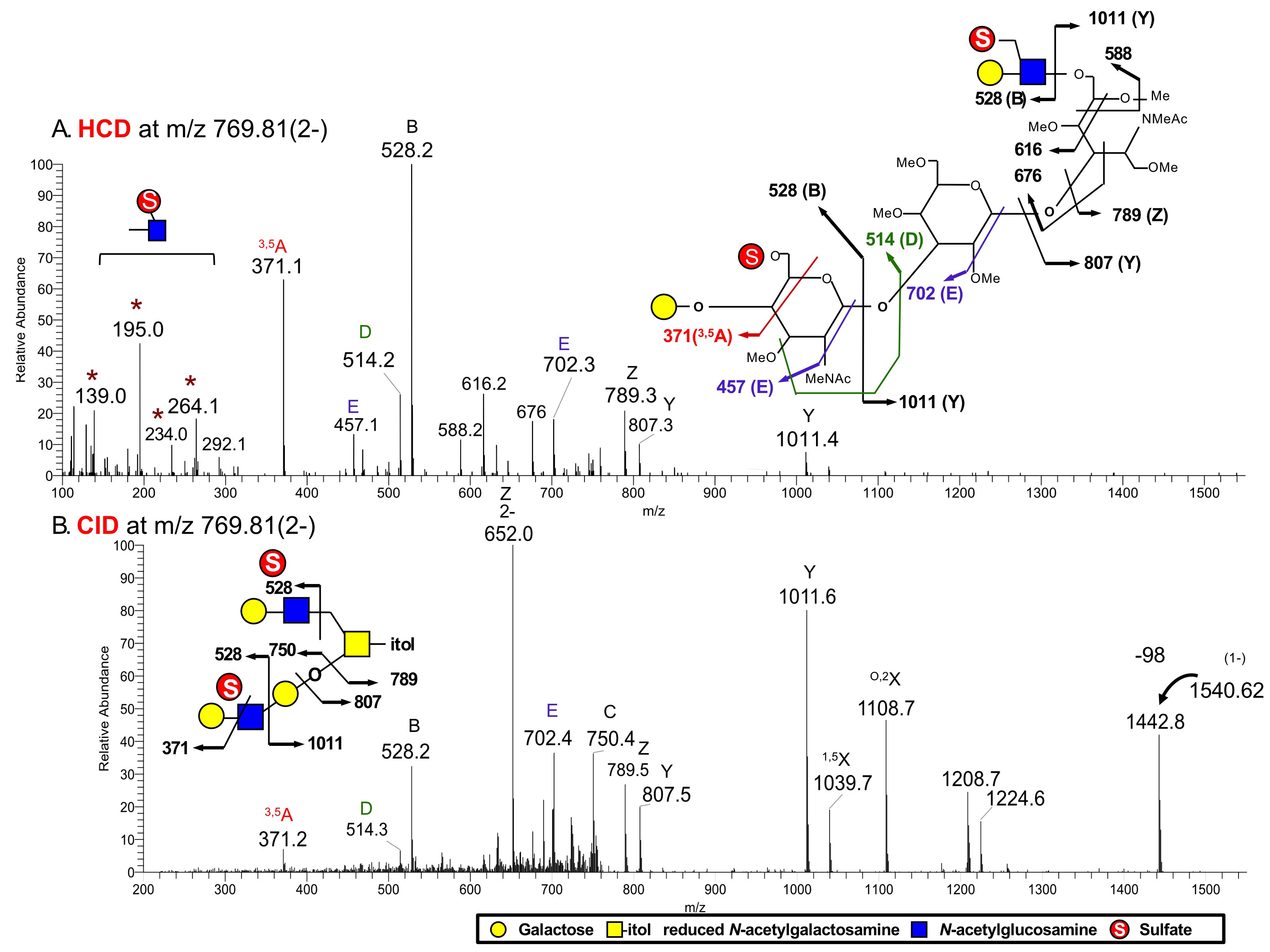
Figure 4. HCD-MS/MS and CID-MS/MS spectra of di-sulfated O-glycan corresponding to 2SO3Hex3HexNAc3-itol at m/z 769.81, doubly charged [M-2H]2-
Table 1. A selection of diagnostic fragment ions afforded by HCD-MS/MS analyses that can be used to assess the presence and relative abundance of some of the most commonly found sulfated glycotopes. The MS2 ions at m/z 528, 702, 889, 1063 can be further subjected to MS3 to produce those diagnostic ions at < m/z 400 that are informative of the precise location of sulfate on either the Gal or GlcNAc residue. The signal at m/z 97 is highly abundant, ubiquitous and useful to confirm presence of sulfate but itself is not informative of the particular sulfated glycotopes. It can be excluded from the mass range of LC-MS2 data acquisition to better detect the other much lower abundant ions.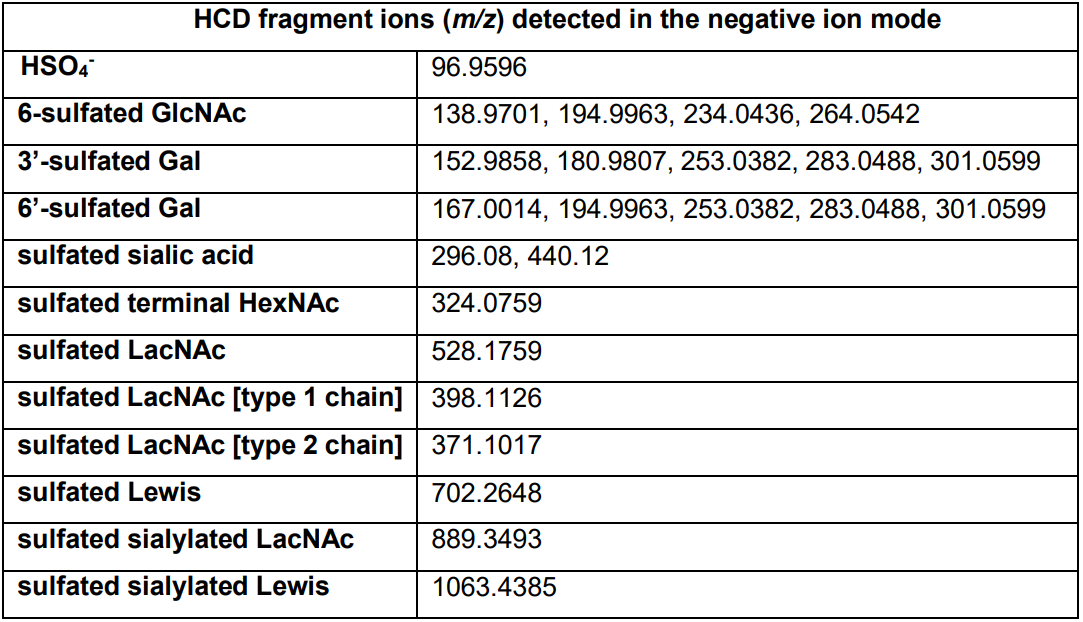
- HCD-MS/MS analysis
- A strong B ion at m/z 528 and related E ion at m/z 457 indicate the existence of sulfated LacNAc unit.
- 3,5A ion at m/z 371 and D ion at m/z 514 indicate the existence of sulfated type 2 LacNAc.
- Low mass ions at m/z 139, 195, 234, 264 indicate that both sulfate groups are located on the 6-carbon position of HexNAc.
- The ions at m/z 588/616/676 are cleavage ions at HexNAc-itol, indicating that the sulfated LacNAc unit is carried on the 6-arm of HexNAc-itol, which are informative. This glycotope is further confirmed by Y ion at m/z 807 and Z ion at m/z 789.
- CID-MS/MS analysis
- Those fragment ions shown in CID MS2 were also observed in the HCD MS2 data except i) ion at m/z 1442, corresponding to elimination of sulfate from precursor ion; ii) Z ion at m/z 652, which is doubly charged and corresponds to the elimination of terminal hexose residue; iii) ions at m/z 1039 and 1108 are X ions at HexNAc residue; iv) ions at m/z 1224 and 1208, derived from loss of hexose from the ion at m/z 1442 and concerted elimination of sulfate and hexose from precursor ion at m/z 1540, respectively (Yu et al., 2006).
- Informative low mass fragment ions, which would indicate the location of sulfate, are lost in the CID-MS/MS data due to cut-off.
- Readers are referred to Figure 4 in the published work (Cheng et al., 2015) for the characteristic MS2 fragment ions afforded by HCD-MS/MS analyses that are useful to identify the location of sulfate. These characteristic fragment ions were shown not only in HCD-MS/MS analyses of permethylated sulfated O-glycans, but also N-glycans.
- CID-MS3 spectra of permethylated sulfated glycans can afford information about sulfated Gal and sulfated GlcNAc, but not the exact location of sulfate (Figures 3C and 3D in Cheng et al., 2015). This can be further resolved and defined by HCD-MS3 analysis enabled on the Orbitrap Fusion (Figure S3 in Hsiao et al., 2017).
- Currently, singly charged precursor ions for permethylated mono-sulfated N-glycans and O-glycans at over m/z 2000 in the negative ion mode cannot be efficiently isolated in the quadrupole or trap of Orbitrap MS systems for MS/MS analysis.
- HCD-MS/MS analysis
- In-house GlyPick computation tool
- Additional mapping of the sulfated glycotopes based on their respective MS2 diagnostic ions (Figure 5) can be performed using the in-house developed software, GlyPick. The representative figure describing the user interface for GlyPick has been shown as Figure S4 in Hsiao et al. (2017).
- Parameters in GlyPick for input data of permethylated sulfated glycans
- Set up the mass tolerance at ± 5 ppm and choose Orbitrap as instrument type for the target HCD-MS2 to be extracted from the acquired data sets, with their absolute and relative intensity threshold set at above 100 and 1%.
- Choose negative ion mode.
- Select the known diagnostic fragment ions from a built-in list as target ions. Additional ions not in the list can be user-input.
- Choose “Reduced” as reducing end when analyzing permethylated reduced glycans.
- Modify other parameters depending on different samples such as mono-sulfated or di-sulfated; N-glycans or O-glycans.
- MS3 setting will be used when pd-MS3 acquisition is additionally performed.
- The summed ion intensity for each of the extracted target ions will be computed and the results output in CSV format. These can be further calculated as percentage total of all selected MS2 ions, as a convenient way of normalization.
Acknowledgments
This LC-MS/MS experimental workflow for sulfoglycomics was adapted from two published works (Cheng et al., 2015; Yu et al., 2018) while additional glycotope-centric data mining with GlyPick was developed by Hsiao et al. as reported in Hsiao et al., 2017. MS data was acquired by LTQ-Orbitrap at the Academia Sinica Common Mass Spectrometry Facilities for Proteomics and Protein Modification Analysis located at the Institute of Biological Chemistry, Academia Sinica, supported by Academia Sinica Core Facility and Innovative Instrument Project (AS-CFII-108-107). These works were supported by Academia Sinica and Taiwan Ministry of Science and Technology (MoST) grants to KKH.
Competing interests
The authors declare no conflict of interest.
References
- Cheng, C. W., Chou, C. C., Hsieh, H. W., Tu, Z., Lin, C. H., Nycholat, C., Fukuda, M. and Khoo, K. H. (2015). Efficient mapping of sulfated glycotopes by negative ion mode nanoLC-MS/MS-Based sulfoglycomic analysis of permethylated glycans. Anal Chem 87(12): 6380-6388.
- Cheng, P. F., Snovida, S., Ho, M. Y., Cheng, C. W., Wu, A. M. and Khoo, K. H. (2013). Increasing the depth of mass spectrometry-based glycomic coverage by additional dimensions of sulfoglycomics and target analysis of permethylated glycans. Anal Bioanal Chem 405(21): 6683-6695.
- Hsiao, C. T., Wang, P. W., Chang, H. C., Chen, Y. Y., Wang, S. H., Chern, Y. and Khoo, K. H. (2017). Advancing a High Throughput Glycotope-centric Glycomics Workflow Based on nanoLC-MS(2)-product Dependent-MS(3) Analysis of Permethylated Glycans. Mol Cell Proteomics 16(12): 2268-2280.
- Patnode, M. L., Cheng, C. W., Chou, C. C., Singer, M. S., Elin, M. S., Uchimura, K., Crocker, P. R., Khoo, K. H. and Rosen, S. D. (2013). Galactose 6-O-sulfotransferases are not required for the generation of Siglec-F ligands in leukocytes or lung tissue. J Biol Chem 288(37): 26533-26545.
- Yu, S. Y., Hsiao, C. T., Izawa, M., Yusa, A., Ishida, H., Nakamura, S., Yagi, H., Kannagi, R. and Khoo, K. H. (2018). Distinct substrate specificities of human GlcNAc-6-sulfotransferases revealed by mass spectrometry-based sulfoglycomic analysis. J Biol Chem 293(39): 15163-15177.
- Yu, S. Y., Snovida, S. and Khoo, K. H. (2006). Permethylation and microfractionation of sulfated glycans for MS analysis. Bio-protocol 10(10): e3617.
- Yu, S. Y., Wu, S. W. and Khoo, K. H. (2006). Distinctive characteristics of MALDI-Q/TOF and TOF/TOF tandem mass spectrometry for sequencing of permethylated complex type N-glycans. Glycoconj J 23(5-6): 355-369.
Article Information
Copyright
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Yu, S., Cheng, C. and Khoo, K. (2020). Negative Ion Mode nanoLC-ESI-MS/MS Analyses of Permethylated Sulfated Glycans. Bio-protocol 10(10): e3618. DOI: 10.21769/BioProtoc.3618.
- Yu, S. Y., Hsiao, C. T., Izawa, M., Yusa, A., Ishida, H., Nakamura, S., Yagi, H., Kannagi, R. and Khoo, K. H. (2018). Distinct substrate specificities of human GlcNAc-6-sulfotransferases revealed by mass spectrometry-based sulfoglycomic analysis. J Biol Chem 293(39): 15163-15177.
Category
Biochemistry > Carbohydrate > Polysaccharide
Biochemistry > Carbohydrate > Glycoprotein
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link