- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Determination of Chromatin Accessibility in Drosophila Midgut Enterocytes by in situ 5mC Labeling
Published: Vol 9, Iss 22, Nov 20, 2019 DOI: 10.21769/BioProtoc.3435 Views: 4437
Reviewed by: Alexandros AlexandratosTzvetina BrumbarovaAbhijit Kale

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Dual-Color Live Imaging of Adult Muscle Stem Cells in the Embryonic Tissues of Drosophila melanogaster
Monika Zmojdzian [...] Rajaguru Aradhya
Feb 5, 2023 1679 Views

Live Imaging of Phagoptosis in ex vivo Drosophila Testis
Diana Kanaan [...] Hila Toledano
Mar 20, 2023 1831 Views

Protocol for Imaging the Same Class IV Neurons at Different Stages of Development
Sonal Shree and Jonathon Howard
Aug 20, 2024 1320 Views
Abstract
Regulation of gene expression involves dynamic changes in chromatin organization, where in many cases open chromatin structure correlates with gene activation. Several methods enable monitoring changes in chromatin accessibility, including ATAC-seq, FAIRE-seq, MNase-seq and DNAse-seq methods, which involve Next-generation-sequencing (NGS). Focusing on the adult Drosophila differentiated gut enterocytes (ECs) we used a sequencing-free method that enables visualizing and semi-quantifying large-scale changes in chromatin structure using in vitro methylation assay with the bacterial CpG Methyltransferase, M. Sssl, that determine chromatin accessibility. In brief, as CpG methylation is minimal in differentiated somatic Drosophila cells, we used the bacterial M. SssI enzyme to methylate CpG dinucleotides in situ depending on their chromatin accessibility. The methylated dinucleotides are detected using 5mCytosine monoclonal antibody and nuclei are visualized microscopically. Thus, the 5mC method enables to monitor large-scale chromatin changes in heterogenic cellular tissue focusing on the cell type of interest and without the need for cell purification or NGS.
Keywords: Chromatin 5-methyl-Cytosine (5mC)Background
The regulation of the differentiated state of cells requires active supervision and is established and maintained by “identity supervisors” (Natoli, 2010; Holmberg and Perlmann, 2012). In part, these supervisors are transcription factors (TFs) that together with chromatin and architectural/scaffold proteins establish and maintain large-scale organization of the differentiated nucleus (Bitman-Lotan and Orian, 2018). In both mammals and Drosophila, the adult gut is a highly dynamic tissue where intestinal stem cells (ISCs) proliferate to self-renew or differentiate to give rise to mature differentiated gut polyploid enterocytes (ECs) (Jiang and Edgar, 2012; Buchon et al., 2013; Guo et al., 2016). In the differentiated ECs, the transcription factor Hey together with the Drosophila type A lamin, Lamin-C, co-regulate enhancers activity and large-scale nuclear organization that prevents the expression of progenitor and non-relevant programs (Flint-Brodsly et al., 2019). Genetic ablation of Hey in young ECs, or due to its decline during aging lead to a failure to express EC programs, including a decrease in LamC protein. Loss of LamC resulted in the ectopic expression of non-relevant gene programs and re-organization of the nucleus. These large-scale changes in chromatin organization can be visualized using the 5mC method as shown in Figures 1A and 1B. Moreover, the 5mC method was also used to monitor changes in chromatin accessibility in other Drosophila tissues such as the ovary upon loss of upSETR, a chromatin regulator (Rincon-Arano et al., 2012). Please note that while the method provides a qualitative approach to address chromatin organization in complex tissues, NGS-based approaches should be considered to confirm and fine map structural changes.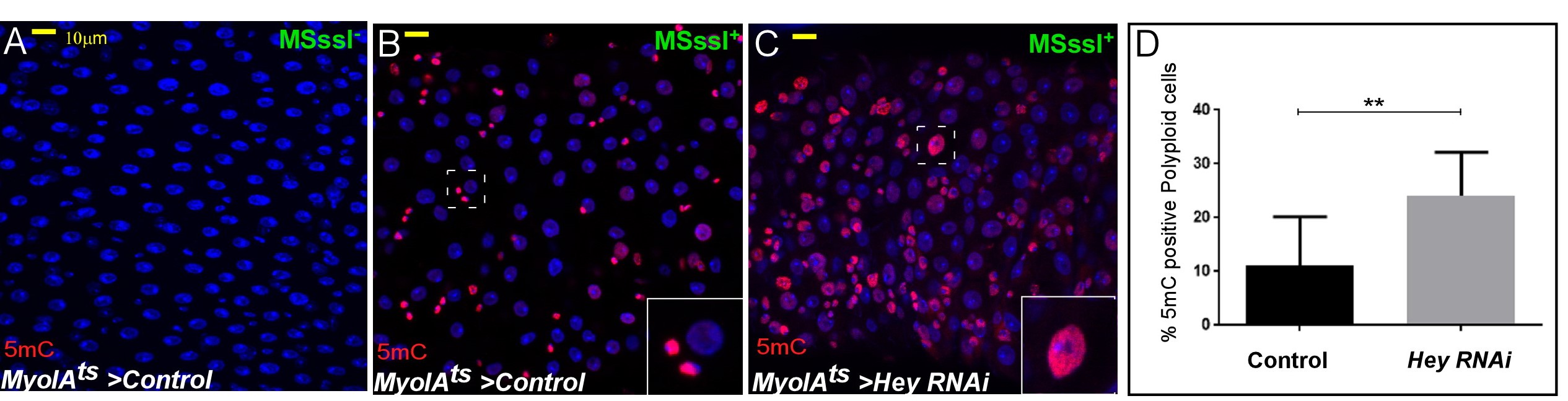
Figure 1. Loss of Hey in ECs results in increased chromatin accessibility in polyploid cells.Confocal images of midguts expressing the indicated transgenes: A. Control midgut stained for 5mC without M. SssL-I enzyme. B. Control midgut with enzyme treatment. C. Midguts where Hey was knockdown in ECs. 5mC methylation is shown in red, DAPI marks DNA, scale bar = 10 μM, dashed squares point to cells shown in the inset. D. Quantification of 5mC positive polyploid cells in similar setting to B and C. ** = P<0.01 (adopted from Flint-Brodsly et al., 2019).
General description of the method: The M. SssI methyltransferase modifies all cytosine residues (C5) within the double-stranded dinucleotide recognition sequence CG, and it has been used for nucleosome mapping from isolated nuclei, during transcription (Fatemi et al., 2005; Bell et al., 2010). As the Drosophila genome has only minimal endogenous 5-cytosine methylation (5mC), we established conditions that enable efficient modification of mCpG dinucleotides by M. SssI methyltransferase depending on chromatin accessibility in Drosophila guts in situ. After a mild fixation and efficient permeabilization of the guts, M. SssI-based CpG methylation is performed at different time points and the DNA is denatured to expose the modified DNA. The resulting methylation is observed using a 5m Cytosine specific monoclonal antibody (Bell et al., 2010), and is low or absent in untreated ECs in wild-type tissues (Not shown). M. SssI-based 5mC methylation is only minimally observed in wildtype polyploid ECs (Figure 1A). However, it dramatically increases in polyploid cells resembling ECs upon loss of Hey (Figures 1B and 1C; Flint-Brodsly et al., 2019). Quantification of the visual signal demonstrated that the 5mC signal was observed in 11% of control ECs, and in 24% of Hey-targeted ECs (Figure 1C, n = 395, 820 respectively, P< 0.01).
The 5mC method described below (Outlines in Figure 2) is simple to use, reproducible, enables focusing on specific cells of interest and can be combined with linage-tracing methods such as G-TRACE (Evans et al., 2009).
Moreover, it does not involve cell purification or NGS, and is semi-quantitative. However, it requires the appropriate controls, including scaling the duration of the methylation reaction, fixation, permeabilization conditions and enzyme adjustments depending on the tissue of choice. Additional semi-quantitative evaluation can be performed by extracting the DNA from treated tissues and using nuclei acid-specific ELISA or DNA-based dot blot to estimate the dynamic range of the 5mC methylation. Moreover, unlike NGS-based methods (Tsompana and Buck, 2014), it does not point to the genomic location of the accessible regions. 
Figure 2. 5mC method flow chart
Materials and Reagents
- Aluminum foil
- Eppendorf tubes
- Pipettes range of 0-10 μl, 10-200 μl, 200-1,000 μl
- Microscope mounting Slides (Sigma-Aldrich, catalog number S8902)
- Cover slips (Thermo Fisher, catalog number 102455)
- Albumin Bovine, Fraction V (MP biomedicals, catalog number: 0216006980)
- CpG Methyltransferase (M. SssI) (Supplied with Buffer and S-adenosylmethionine [SAM])- (New England Bio-labs (NEB), catalog number: M0226L)
- Alexa Fluor 568 goat anti-mouse IgG (H + L) (Invitrogen, catalog number: A11031)
- 5-Methylcytosine (5-mC) antibody (mAb) - clone 33D3 (Active Motif, catalog number: 39649)
- Schneider's Drosophila Medium (Biological Industry, catalog number: 01-150-1A)
- Dulbecco's Phosphate Buffered Saline (DPBS) (10x) (Biological industries, catalog number: 02-023-5A)
- PierceTM 16% Formaldehyde (w/v), Methanol-free (Thermo ScientificTM, catalog number: 28906)
- Heptane (Sigma-Aldrich, catalog number: 246654)
- Triton X-100 (Sigma-Aldrich, catalog number: X100)
- 32% Hydrochloric acid (Bio-Lab, catalog number: 084605)
- Borax anhydrous (Sigma-Aldrich, catalog number: 71997)
- DAPI-for nucleic acid staining (Sigma-Aldrich, catalog number: D9542-1MG)
- Fluoromount-G (Southern Biotech, catalog number: 0100-01)
- HCl
- Fix-I solution (1 ml) (see Recipes)
- PBX (see Recipes)
- PAT (see Recipes)
- Pre-reaction Buffer (1 ml) (see Recipes)
- M. SssI Reaction Buffer-100 μl (25 U) (see Recipes)
- Fix-II solution (1 ml) (see Recipes)
- PBT (see Recipes)
Equipment
- Incubator for rearing Drosophila at consistent temperature of 29 °C
- Stereomicroscope with light source for dissection (Leica Microsystems, model: MZ75)
- Dumont tweezers #5 Biology 0.05 x 0.02 mm Inox (magnetic, FST11252-20)
- Watch Glasses square for dissection (Carolina, catalog number: 742300)
- Analog Orbital Shaker (ELMI S-3.02 10 L)
- Confocal microscope (Zeiss, model: LSM-700)
Procedure
- Flies collection
Drosophila flies are reared at 22 °C on standard medium. To prevent gender differences, collect only female flies at relevant age. For collecting young adults, we used 3-10 days old flies that were transferred to 29 °C for two days for activating the GAL4/Gal80 system and UAS-RNAi for gene knockdown. For more details regarding the GAL4/GAL80 system, see Salmeron et al. (1990) and Brand and Perrimon (1993). - 5mC methylation assay
- Dissect guts in watch glasses squares filled with cold Schneider medium. Transfer dissected guts to an Eppendorf tube containing 0.5 ml Schneider medium. Eppendorf tubes with dissected guts are kept on ice. Collect 8-10 guts per Eppendorf tube. For detailed gut dissection, see an excellent midgut protocol (Nawrot-Esposito et al., 2017).
- Remove Schneider media and add 1 ml cold DPBS to wash dissected guts (replace media only without incubation time).
- Fix: Replace DPBS with 1 ml of freshly made "Fix-I solution" (Recipe 1), this will form two phases of liquids: the upper phase is Heptane and lower phase is the formaldehyde/DPBS solution at the bottom (See Figure 3 “Fix”). Rotate for 20 min on an orbital shaker at RT.
Note: Over-crosslinking can cause reduced diffusion and accessibility of the enzyme to the nuclei.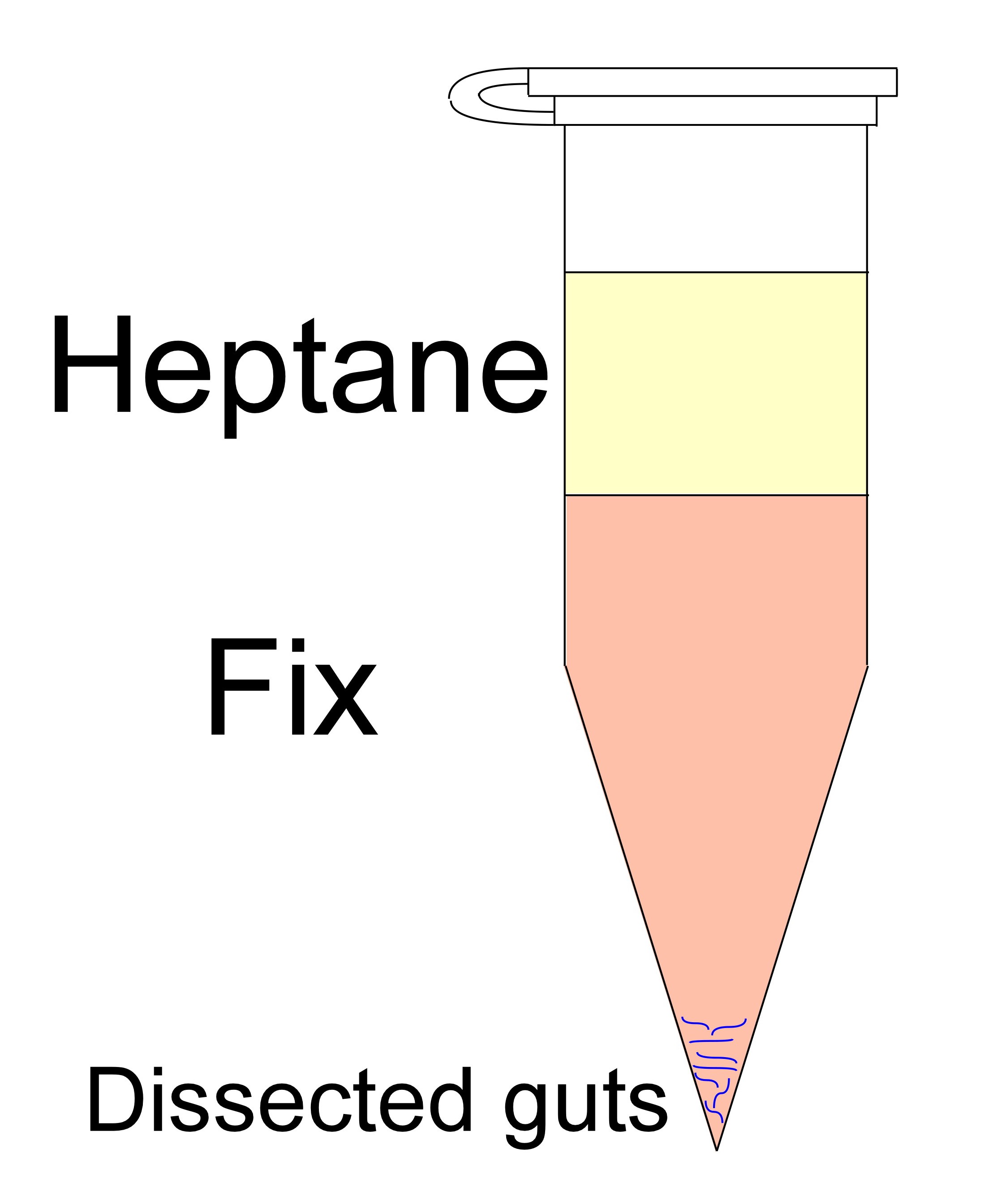
Figure 3. Typical Eppendorf tube after formation of the two phases - Gently remove all the liquid solutions (the 2 phases) leaving only the dissected tissue at the bottom. Be sure to remove all heptane, also from the cap.
- Wash guts three times with 1 ml DPBS, in each wash rotate the guts for 15 min at RT.
- Permeabilization: Permeabilize the tissue by incubating with 500 µl of 0.5% Triton X-100 in DPBS for 1h on an orbital shaker in Eppendorf tubes, at RT (we recommend evaluating different permeabilization conditions [e.g., 1 h, 2 h and triton concentrations (0.5%, 0.6%, 0.7%)]).
- Gently remove Triton solution, and wash for three times with DPBS, in each wash rotate the guts for 15 min at RT.
- Block in 1 ml "PAT solution" (Recipe 2) overnight at 4 °C on an orbital shaker.
- Wash twice with freshly prepared 250 μl of "Pre-reaction Buffer "(Recipe 3), 15 min each wash.
- Methylation reaction: Gently remove "Pre-Reaction Buffer” and resuspend in 50 µl of M. SssI reaction buffer (Recipe 4) and incubate for 1 h at 25 °C, on an orbital shaker.
Important: Titer the reaction conditions time (e.g., 1 h, 5 h, 24 h) and enzyme amount (25 U, 50 U) for optimal results. - Termination: Add 1 ml of 2 N HCl and rotate on an orbital shaker at room temperature for 30 min. This step stops the M. SssI reaction and denatures the DNA.
- Remove HCl solution and neutralize with 0.5 ml of freshly prepared 100 mM Borax (dissolved in water) and rotate on an orbital shaker for 5 min at RT.
- Remove Borax and wash twice with 1 ml DPBS, rotate the guts for 15 min at RT each wash.
- Fix with freshly prepared "Fix-II solution" (Recipe 5) and rotate for 10 min.
- Remove Fix-II and wash twice with 1 ml "PBX solution" (Recipe 6), rotate the guts for 15 min at RT each wash.
Note: Evaluate different time points (10, 15, 20 min) as over-crosslinking could affect antibody penetration. - Remove PBX and block with 1 ml "PAT solution" (Recipe 2), for 30 min rotating at RT.
- Remove blocking solution and add diluted anti-5mC primary antibody in "PAT solution" (Recipe 2), to a concentration of 1 μg/ml (1:1,000). Incubate overnight at 4 °C on an Orbital shaker.
- Remove primary antibody and wash three times with "PBT solution" (Recipe 7), rotating for 15 min each wash at RT.
- Remove PBT and incubate in diluted Secondary Fluorescent anti-mouse (1:1,000) together with DAPI (1:1,000) (for DNA staining) in "PAT solution" (Recipe 2), cover the samples with aluminum foil or in a light protected box, and rotate on orbital shaker for 1 h in the dark.
- Remove Secondary antibody and DAPI solution and wash three times with "PBT solution" (recipe 7), 15 min each wash while rotating in light protected environment (box or aluminum foil) at RT.
- Wash twice with 1 ml DPBS, rotate the guts for 15 min at RT each wash.
- Mount in a Fluoromount-G (25 ml) on slide and keep at 4 °C until confocal microscopic analysis.
- Visualize under a confocal microscope, signal intensity in cells can be quantified. For quantification we suggest counting several hundreds of cells derived from different biological repeats, and in each scoring 5mC positive polyploid cells.
Recipes
Prepare each solution fresh and keep no longer than a week at 4 °C or on Ice.
- Fix-I solution (1 ml)
75 μl 16% EM-grade formaldehyde
12.5 μl 10x DPBS
112.5 μl distilled water
600 μl Heptane - PAT
1% BSA in PBX - Pre-reaction Buffer (1 ml)
899.5 μl distilled water
100 μl 10x NEB buffer (New England Biolab MsslI buffer)
0.5 μl SAM (S-adenosyl methionine from NEB, supplied with the enzyme) - M. SssI Reaction Buffer-100 μl (25 U)
6.25 μl M. SssI
83.25 μl distilled water
10 μl 10x NEB
0.5 μl SAM - Fix-II solution (1 ml)
50 μl 16% EM-grade formaldehyde
150 μl 1x DPBS - PBX
0.1% Triton X-100 in 1x DPBS - PBT
0.1% BSA in PBX
Acknowledgments
This original protocol was used in Rincon-Arano et al. (2012). The research was supported by Israel Science Foundation (ISF) (Grant 739/15), and the Rappaport Institute for Research in the Medical Sciences and Flinkman-Marandi cancer research grant to AO.
Competing interests
We declare that there is no financial or non-financial conflict of interest, or competing interests related to this work to any of the authors.
References
- Bell, O., Schwaiger, M., Oakeley, E. J., Lienert, F., Beisel, C., Stadler, M. B. and Schubeler, D. (2010). Accessibility of the Drosophila genome discriminates PcG repression, H4K16 acetylation and replication timing. Nat Struct Mol Biol 17(7): 894-900.
- Bitman-Lotan, E. and Orian, A. (2018). Chromatin, Nuclear Lamins, and maintenance of the differentiated identity. Curr Opin System Biol 11: 1-8.
- Brand, A. H. and Perrimon, N. (1993). Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118(2): 401-415.
- Buchon, N., Osman, D., David, F. P., Fang, H. Y., Boquete, J. P., Deplancke, B. and Lemaitre, B. (2013). Morphological and molecular characterization of adult midgut compart-mentalization in Drosophila. Cell Rep 3(5): 1725-1738.
- Evans, C. J., Olson, J. M., Ngo, K. T., Kim, E., Lee, N. E., Kuoy, E, Patananan, A. N., Sitz, D., Tran, P., Do, M. T., Yackle, K., Cespedes, A., Hartenstein, V., Call, G. B. and Banerjee, U. (2009). G-TRACE: Rapid Gal4-based cell lineage analysis in Drosophila. Nat Methods 6(8): 603-605.
- Fatemi, M., Pao, M. M., Jeong, S., Gal-Yam, E. N., Egger G, Weisenberger, D. J. and Jones, P. A. (2005). Footprinting of mammalian promoters: use of a CpG DNA methyltransferase revealing nucleosome positions at a single molecule level. Nucleic Acids Res 33(20): e176.
- Flint-Brodsly, N., Bitman-Lotan, E., Boico, O., Shafat, A., Monastirioti, M., Gessler, M., Delidakis, C., Rincon-Arano, H. and Orian, A. (2019). The transcription factor Hey and nuclear lamins specify and maintain cell identity. Elife 8. pii: e44745.
- Guo, Z., Lucchetta, E., Rafel, N. and Ohlstein, B. (2016). Maintenance of the adult Drosophila intestine: All roads lead to homeostasis. Curr Opin Genet Dev 40: 81-86.
- Holmberg, J. and Perlmann, T. (2012). Maintaining differentiated cellular identity. Nat Rev Genet 13: 429-439.
- Jiang, H. and Edgar, B. A. (2012). Intestinal stem cell function in Drosophila and mice. Curr Opin Genet Dev 22(4): 354-360.
- Nawrot-Esposito, M., Loudhaief, R. and Gallet, A. (2017). Protease activity assay in fly intestines. Bio-protocol 7(18): e2560.
- Natoli, G. (2010). Maintaining cell identity through global control of genomic organization. Immunity 33: 12-24.
- Rincon-Arano, H., Halow, J., Delrow, J. J., Parkhurst, S. M., and Groudine, M. (2012). UpSET recruits HDAC complexes and restricts chromatin accessibility and acetylation at promoter regions. Cell 151(6): 1214-1228.
- Salmeron-Jr, J. M., Leuther, K. K. and Johnston, S. A. (1990). GAL4 mutations that separate the transcriptional activation and GAL80-interactive functions of the yeast GAL4 protein. Genetics 125(1): 21-27.
- Tsompana, M. and Buck, M. J. (2014) Chromatin accessibility: a window into the genome. Epigenetics Chromatin 7(1): 33.
Article Information
Copyright
Bitman-Lotan et al. This article is distributed under the terms of the Creative Commons Attribution License (CC BY 4.0).
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Bitman-Lotan, E., Rincon-Arano, H., Raz, G. and Orian, A. (2019). Determination of Chromatin Accessibility in Drosophila Midgut Enterocytes by in situ 5mC Labeling. Bio-protocol 9(22): e3435. DOI: 10.21769/BioProtoc.3435.
- Flint-Brodsly, N., Bitman-Lotan, E., Boico, O., Shafat, A., Monastirioti, M., Gessler, M., Delidakis, C., Rincon-Arano, H. and Orian, A. (2019). The transcription factor Hey and nuclear lamins specify and maintain cell identity. Elife 8. pii: e44745.
Category
Molecular Biology > DNA > Chromatin accessibility
Cell Biology > Cell imaging > Confocal microscopy
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link












