- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Heterochronic Phenotype Analysis of Hypodermal Seam Cells in Caenorhabditis elegans
Published: Vol 9, Iss 1, Jan 5, 2019 DOI: 10.21769/BioProtoc.3132 Views: 6534
Reviewed by: Manish ChamoliSanjib GuhaAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Auxin-mediated Protein Degradation in Caenorhabditis elegans
Michael A. Q. Martinez and David Q. Matus
Apr 20, 2020 5743 Views

Labelling of Active Transcription Sites with Argonaute NRDE-3—Image Active Transcription Sites in vivo in Caenorhabditis elegans
Antoine Barrière and Vincent Bertrand
Jun 5, 2022 2683 Views

Simple and Rapid Model to Generate Differentiated Endometrial Floating Organoids
Adriana Bajetto [...] Tullio Florio
Feb 5, 2026 92 Views
Abstract
Heterochrony refers to changes in the timing of developmental events, and it is precisely regulated in the organisms by the heterochronic genes such as C. elegans lin-4 and let-7. Mutations in these genes cause precocious or retarded development of certain cell lineages. With well-defined cell lineages, C. elegans is one of the best model systems to study heterochronic genes, since the subtle changes in the development of cell lineages can be easily identified. Among the different cell types in C. elegans, hypodermal seam cells and their lineages are well known to be maintained by lin-14, whose expression level is regulated by two miRNA genes, lin-4 and let-7, at the larval stages. Therefore, analyzing the heterochronic phenotype of hypodermal seam cells in C. elegans could yield detailed insights into the status of the miRNA pathway. Here we describe the assay protocol to analyze the heterochronic phenotypes of C. elegans hypodermal seam cells, which can be used as a reliable method to study the miRNA pathway.
Keywords: C. elegansBackground
Caenorhabditis elegans is a transparent nematode which is found in the soil. It was introduced as a new model organism by Sydney Brenner in the 1960s to study neural development. Since then, it has been extensively studied because of the simple anatomy, the easy cultivation, and the rapid growth. The reproductive life cycle of C. elegans, which takes only three days, consists of the embryonic stage, four larval stages (L1-L4), and the adult stage. After 14 h of embryogenesis, C. elegans grows in size during the four larval stages which are divided by each molt, then it reaches the adult stage which can produce the next generation. In unfavorable conditions, C. elegans larvae can choose the alternative developmental pathway, so-called “dauer”, which can survive a few months in the adverse conditions. When the environment becomes favorable, C. elegans exits the dauer stage and develops into the L4 stage.
In C. elegans, most hypodermal seam cells have the characteristic of stem cells. In each molt, they divide into two daughter cells; the anterior daughter cell fuses with hyp7, which is the major hypodermis, and the posterior daughter cell continues to divide like the stem cells until they terminally differentiate at the L4 stages. Thus, ten hypodermal seam cells at L1 stage generate sixteen hypodermal seam cells at the end of the L4 stage in each side (Figure 1) (Altun and Hall, 2009).
Functionally, hypodermal seam cells are important for the formation of stage-specific cuticles that are composed of various collagen proteins (Thein et al., 2003). They also produce cuticular alae, which are the protruding ridges extending longitudinally along the two sides of the animal over the seam cells (Singh and Sulston, 1978). Alae are produced only in the L1 stage, dauer larva, and adult. Therefore, adult alae are commonly used as an indicator which shows terminal differentiation of hypodermal seam cells. For example, lin-4 or let-7 loss-of-function mutant, which shows the retarded development of hypodermal seam cells, has the alae defects in adult stages, while lin-14 loss-of-function mutant, which shows the precocious development of hypodermal seam cells, has alae in the L3 stage (Hong et al., 2000).
The cell fate of a hypodermal seam cell is regulated by lin-14, whose expression is high in L1 animals, and decreases by the L2 stage (Ruvkun and Giusto, 1989). The lin-14 expression is regulated by well-characterized miRNAs lin-4 and let-7 (Ambros, 1989; Reinhart et al., 2000). It is known that lin-4 and let-7 bind the 3’ untranslated region of lin-14 and downregulate its expression at the larval stages (Lee et al., 1993, Slack et al., 2000). When the expression level of lin-14 is not decreased in the larval stages, hypodermal seam cells abnormally proliferate, generating retarded phenotypes in lin-14 gain-of-function mutants and lin-4 loss-of-function mutants. 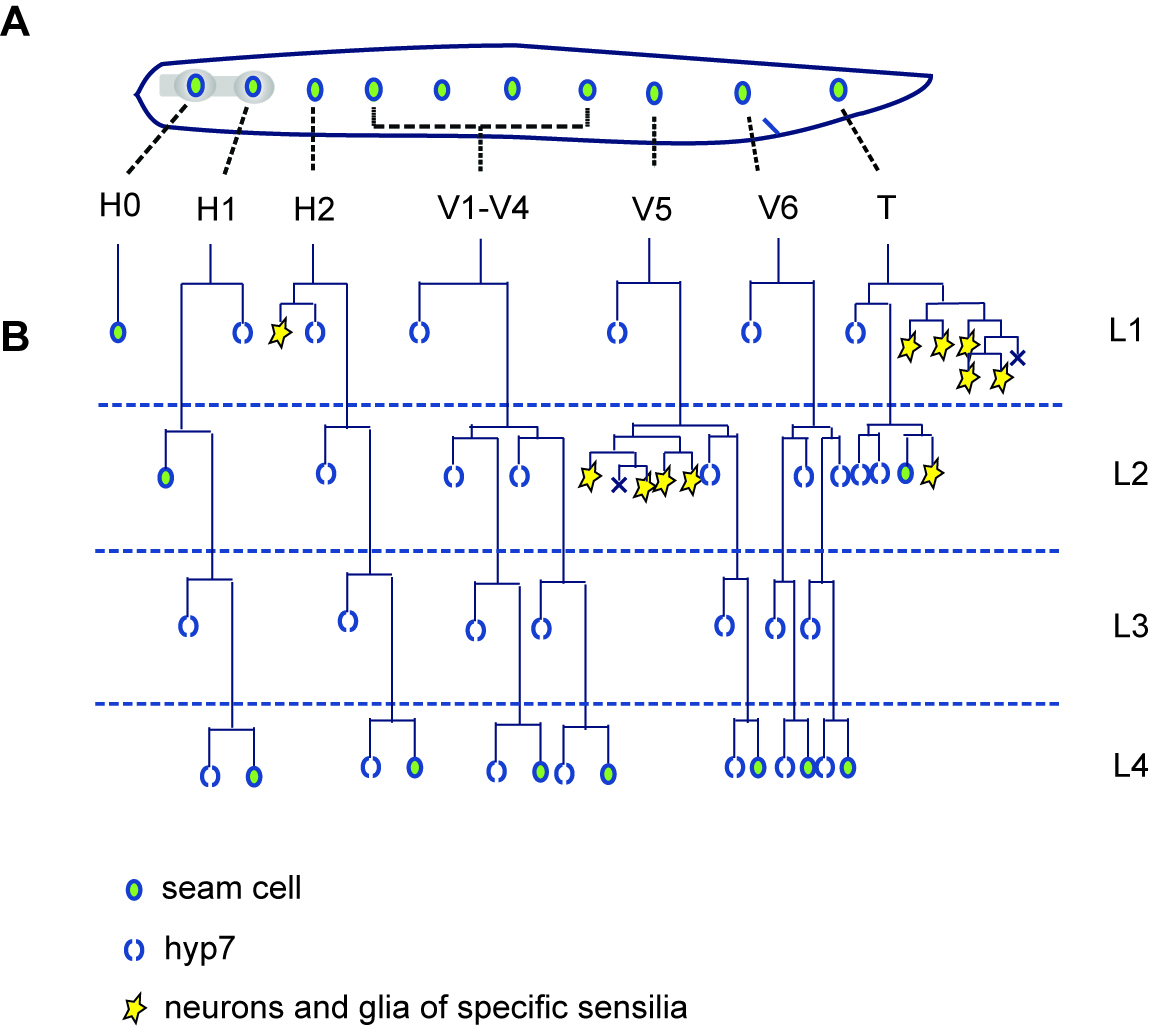
Figure 1. Hypodermal seam cell lineage in C. elegans. A. Ten hypodermal seam cells at the L1 stage. These cells generate major hypodermis hyp7, hypodermal seam cells, neurons and glia throughout the development. B. Cell division patterns of hypodermal seam cells in each molt. Most of the hypodermal seam cells generate one terminally differentiated daughter cell and one stem-cell-like seam cell in each division. By the L4 stage, sixteen of hypodermal seam cells are generated in each lateral side.
LIN-14 is a transcription factor that regulates its target gene expressions. One of the target genes of LIN-14 transcription factor is a cell cycle inhibitor cki-1. Inactivation of cki-1. results in the division of the vulva precursor cell (VPC) during the L2 stage. Therefore, decreasing lin-14 expression via lin-4 miRNA is important for the development of vulva as well as the hypodermal seam cell.
miRNAs are small, non-coding RNAs that are produced by a series of RNA-processing steps from their precursors known as pri-miRNAs. The pri-miRNAs are generated by transcription via RNA polymerase II (Bartel, 2004; Lee et al., 2004) and then cleaved by the RNase III endonuclease Drosha to produce pre-miRNAs in the nucleus (Bracht et al., 2004; Lee et al., 2002). These pre-miRNAs are then transported into the cytoplasm by Exportin-5 (Yi et al., 2003). In the cytoplasm, the pre-miRNA is further processed by Dicer to produce the mature 20-25 nt miRNA (Bernstein et al., 2001; Grishok et al., 2001). The mature miRNA functions as a guide to recruit RNA-induced silencing complex (RISC), which is composed of the miRNA:mRNA duplex, Argonaute, and other proteins (Hammond et al., 2001; Carmell et al., 2002; Caudy et al., 2002; Mourelatos et al., 2002; Caudy et al., 2003). In the RISC complex, Argonaute cleaves the target mRNA when it is activated. In C. elegans, there are about 24 Argonaute proteins (Bartel, 2004; Carmel et al., 2002). Among them, ALG-1 and AGL-2 are appeared to be important for downregulating of lin-4 and let-7 targets, because alg-1 and alg-2 mutants share the phenotypes with lin-4 and let-7 mutants. (Grishok et al., 2001).
Here we describe the assay protocols to analyze the heterochronic phenotypes of hypodermal seam cells in C. elegans. These assays are useful for the analysis of the miRNA pathway by taking advantage of the fact that hypodermal seam cell fate is regulated by the well-characterized miRNAs, lin-4 and let-7 (Zhang et al., 2018).
Materials and Reagents
- 60 mm Petri dishes (Fisher Scientific, FisherbrandTM, catalog number: AS4051)
- 100 mm Petri dishes (Fisher Scientific, FisherbrandTM, catalog number: FB0875712)
- Syringe Filter Unit, 0.22 μm (Millipore Sigma, Millex®-GV, catalog number: SLGV033RS)
- Frosted microscope slides 25 x 75 x 1.0 mm (Fisher Scientific, FisherbrandTM, catalog number: 12-552-3)
- Microscopic cover glass 18 x 18 mm (Fisher Scientific, FisherbrandTM, catalog number: 12-542A)
- C. elegans wild type: N2 Bristol strain from C. elegans Genetic Center
- JR672 wIs54 [Pscm::gfp] V; wIs54 is the integration allele of the seam cell-specific transcriptional GFP reporter that is expressed in all of the seam cells at all developmental stages (Terns et al., 1997; Koh and Rothman, 2001)
- E. coli OP50-1: streptomycin resistant strain from C. elegans Genetic Center
- Levamisole (Sigma-Aldrich, catalog number: L9756)
- NaCl (Fisher Scientific, Fisherbrand, catalog number: S271, CAS 7647-14-5)
- Agar, Bacteriological, Ultrapure (Thermo Scientific, catalog number: J10906, CAS 9002-18-0)
- Peptone (BD Bioscience, BD BactoTM, catalog number: 211677)
- Calcium Chloride hexahydrate (Sigma-Aldrich, catalog number: 442909)
- Magnesium sulfate (Sigma-Aldrich, catalog number: 208094)
- Potassium phosphate dibasic (Sigma-Aldrich, catalog number: P3786)
- Potassium phosphate monobasic (Sigma-Aldrich, catalog number: P0662)
- Cholesterol (Sigma-Aldrich, catalog number: C75209)
- Pure alcohol 200 proof (Pharmco products, catalog number: 111000200)
- Tryptone (BD Bioscience, BD BactoTM, catalog number: 211705)
- Yeast Extract (BD Bioscience, BD BactoTM, catalog number: 212750)
- Nail polish
- Nematode Growth Media (NGM) Agar (see Recipes)
- 1 M CaCl2 (see Recipes)
- 0.5 M MgSO4 (see Recipes)
- 1 M Potassium phosphate (pH 6) (see Recipes)
- 5 mg/ml cholesterol (see Recipes)
- LB agar (see Recipes)
- LB (see Recipes)
- OP50-1 culture (see Recipes)
Equipment
- Amsco® Century SV-120 Scientific Prevacuum Sterilizer (STERIS)
- 4 L flask
- Stir bar
- Stir plate
- PourBoy® 4 Sterile Media Dispenser (TritechTM Research)
- Home-made cell spreader (made with glass Pasteur pipets by bent by heating, spreader size is less than 1 inch.)
- 37 °C incubator (VWR, model: 1535)
- Incubator Shaker (Eppendorf, New Brunswick Scientific, model: I2500, catalog number: M1284-0000)
- 20 °C incubator for C. elegans culture (Intellus control system) (PERCIVAL, model: I-36NL)
- A home-made worm picker with a 5¾ glass Pasteur pipet (Fisher Scientific, FisherbrandTM, catalog number: 13-678-6A) and a platinum wire (Scientific Instrument Services, Inc., catalog number: W414)
- Fluorescent Stereo Microscope (Leica Microsystems, model: Leica M165FC) with Leica PLANAPO 2.0x objective lens
- Confocal laser scanning microscopy platform (Leica Microsystems, model: Leica TCS SP8) with Leica CTR6500 electronics box
Software
- GraphPad Prism 7.0a (GraphPad Software, Inc., www.graphpad.com)
- Leica Application Suite Advanced Fluorescence (Leica Microsystems)
Procedure
- Synchronize the populations and grow the worms (Figure 2A)
- Transfer twenty one-day-old adult worms, which have approximately ten eggs in the uterus, onto 60 mm NGM plates seeded with E. coli (OP50-1).
Note: If your mutant of interest has smaller brood size or egg-laying defects, you may need to transfer more worms onto each plate. - Make 5 replicates of plates per strain.
- Let them lay eggs for two hours. More than fifty eggs per plate are needed. If you have less than fifty eggs per plate, go back to Step A1 and increase the number of adult worms in each plate.
- Remove the adult worms.
- Let the offspring grow until they reach the L4 stage (~48 h) at 20 °C. The L4 stage can be distinguished by a small white crescent spot in the vulva area as well as the size of the worms (Fielenbach and Antebi, 2008).
- Transfer twenty one-day-old adult worms, which have approximately ten eggs in the uterus, onto 60 mm NGM plates seeded with E. coli (OP50-1).
- Analyzing the vulva phenotypes (Figure 2B)
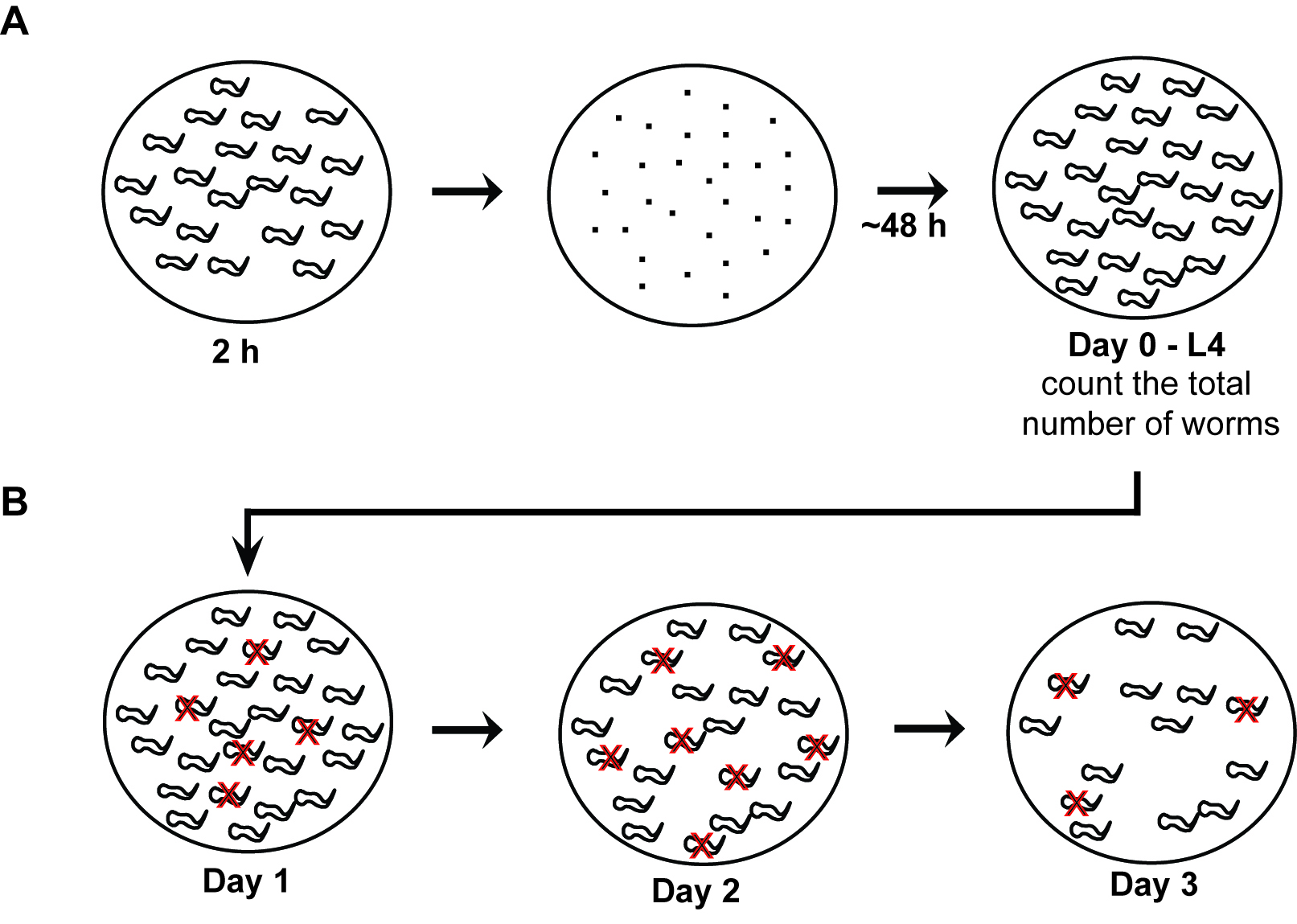
Figure 2. The flow chart of vulva phenotype analysis. A. Synchronization of the worm population. Eggs are collected from young adult worms for two hours, then they are cultured for approximately 48 h until they reach the L4 stage. B. Analysis of vulva phenotype. Count the total number of worms on Day 0. Red X indicates the worms that show vulva phenotype. Count the worms that show the vulva defects and remove them to avoid repeated counting on following days.- Count the total number of L4 worms as soon as they reach the L4 stage. Typically, each plate has approximately one hundred worms.
- After 24 h, censor and remove the worms that show protruded vulva or bagging. Continue with counting and censoring for 3 days.
- Calculate the percentages of vulva defects (Zhang et al., 2018).
Data analysis
- More than 250 worms should be analyzed for each strain, and three independent experiments should be performed.
- Calculate the percentage of worms having vulva defects using the following equation:
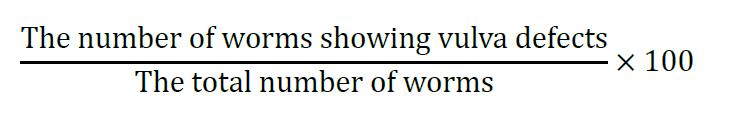
- Enter the value into GraphPad Prism and analyze the mean with SEM and P-value with two-tailed Student’s t-test.
Procedure
- Synchronize the worms as described above.
- Transfer either young adult or the L3 animals to a new plate. Place ten animals per plate. Make five replicates per strain.
- Analyze the alae phenotype. Under the stereomicroscope, focus on the upper side of each worm under the 24x objective lens. You will see the protruding ridge of the alae on the upper side of the worm from the head to the tail. Observe the alae and record whether they are intact, discontinued, or absent in each individual animal. Refer to Slack et al., 2000 for the alae phenotypes. In the adult stage, discontinued or absent alae are abnormal, while the presence of alae is abnormal at the L3 stage.
- Count the number of worms that have intact, discontinued, or absent alae (Zhang et al., 2018).
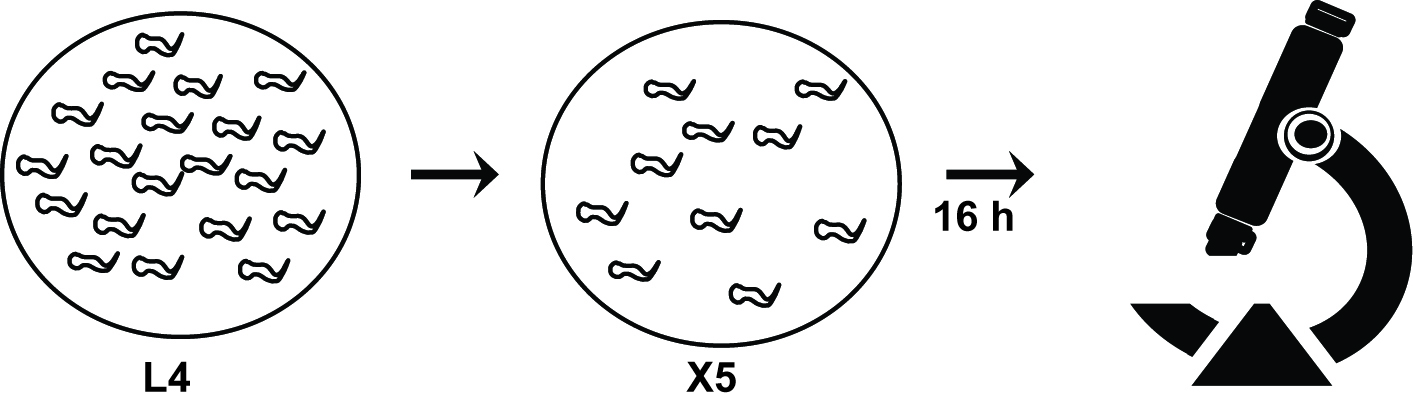
Figure 3. The flow chart of alae phenotype analysis. When synchronized worms reach the specific stage, ten worms are transferred into the new plate and the alae phenotypes are analyzed under the microscope.
Data analysis
- For each strain, at least 50 worms are observed and three independent experiments are performed.
- Calculate the percentage of worms having abnormal alae.
- Means with SEM and P-value with two-tailed Student’s t-test are calculated using GraphPad Prism.
Procedure
- Generating the desired strains expressing Pscm::gfp using JR672 (Figure 4)
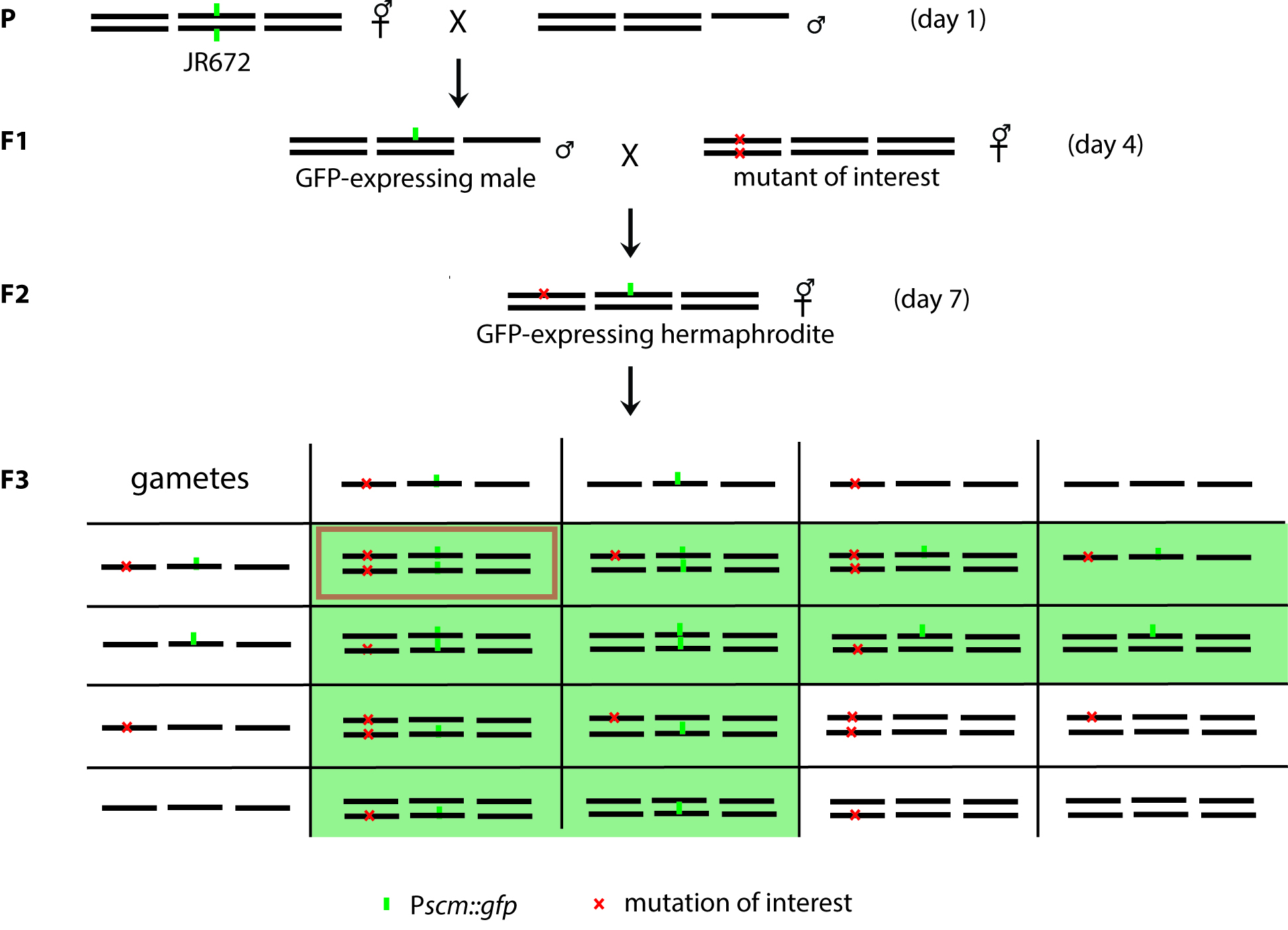
Figure 4. Schematic diagrams of genetic crosses for Pscm::gfp expressing mutants. JR672 is crossed with wild-type males to get the GFP-expressing heterozygote males, which are mated with mutants of interests on Day 4. Then, the GFP-expressing hermaphrodites in the F2 generation are the double heterozygotes. With the Mendelian Segregation, the double heterozygotes from the F2 generation would give rise to double homozygotes with a 1/16 chance, when two mutations are not linked. However, since we select only the GFP-expressing worms, the chance for the double homozygotes is increased to 1/12. The green boxes in F3 indicate the possible genotypes of GFP-expressing worms and the red box indicates the genotype of the double mutant.- Prepare NGM plates for the crosses by seeding 20 μl of OP50 in the middle of 60 mm plates. By making a small bacterial lawn, the chances of worm mating are increased.
- Cross hermaphrodite of the strain JR672 with N2 males. Hermaphrodite to male ratio for the crosses is usually 1:3.
- On Day 3, remove the parent worms to avoid mixing of two generations.
- On Day 4, select F1 males, which are GFP-positive heterozygote to set up a cross with hermaphrodites of your mutant of interest. Keep the hermaphrodite to male ratio 1:3.
- On Day 6, remove the parent worms from the plate. Now, it is expected to have only the F2 generation as larvae or eggs in the plate.
- On Day 7, select the hermaphrodites that express GFP in the hypodermal seam cells. There is a 50% chance to get the GFP-expressing hermaphrodites if the mating is successful. Clone out a couple of GFP-expressing hermaphrodites, one animal per seeded plate.
- On Day 9, remove the parent worm from the plate.
- On Day 10, chose one plate which has a good number of worms. Discard the rest of the plates. From the chosen plate, single out GFP-expressing worms. The chance to get the double mutant is one out of twelve if the mutation is not in the same linkage group as the GFP locus, because only the GFP-expressing worms are selected for analysis. Based on the chance to getting double mutants, select the appropriate number of F3 worms. To increase probability of getting double mutants, pick three to four times number of worms. We usually select forty worms if the mutation of interest is not linked with Pscm::gfp. Note that Pscm::gfp is located on linkage group V.
- On Day 12, genotype F3 worms for the mutation of interest and select the plates which have homozygotes. Check the plates that have homozygous mutants and identify the ones with all F4 animals positive for GFP expression. If desired, GFP homozygotes can be selected by PCR in F3 generation.
- Synchronizing and preparing the worms for microscopy
- To obtain a synchronized population of animals, allow ten to twenty of fertile adult worms to lay eggs for two hours and let them grow until they reach the L4 stages at 20 °C as described in Part I Procedure A.
- Prepare the 2% agarose pads on the imaging slide as described before (Huynh et al., 2018).
- Place ten L4 worms in 7 μl of 10 mM levamisole on the 2% agarose pad, put a coverslip and stabilize the coverslip by painting four corners of coverslip with transparent nail polish.
- Under the 40x objective lens, check the GFP signals. GFP is expressed in the nuclei of hypodermal seam cells. Wild-type worms have sixteen hypodermal seam cells on each side at L4 stage (Figure 5), thus the total number of hypodermal seam cells per worm is 32. By focusing up and down, count the number of hypodermal seam cells on each side. In the tail, the focal planes of right and left hypodermal seam cells are closer than others. Thus it is easier to count the total number of hypodermal seam cells by counting one side of cells from the head to tail and the other side from tail to head.
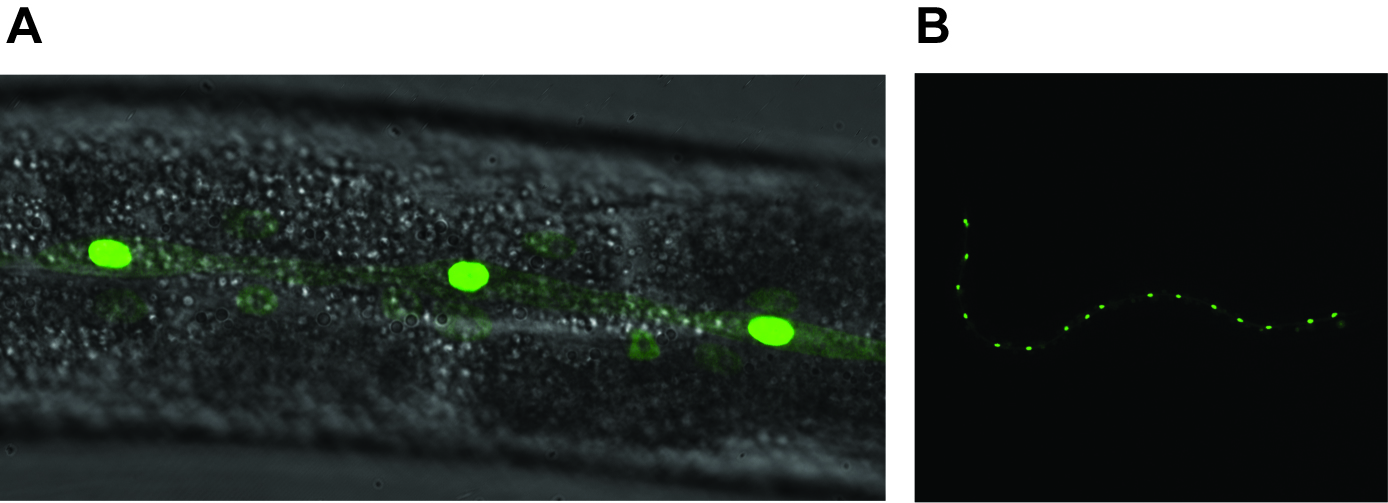
Figure 5. GFP-expression pattern of Pscm::gfp in wild-type C. elegans. A. GFP is expressed in the nuclei of hypodermal seam cells. A 40x objective lens was used in this image. B. Sixteen hypodermal seam cells are shown in wild-type C. elegans under the 10x objective lens. The smear GFP signal is from the other side of hypodermal seam cells (Zhang et al., 2018).
Data analysis
- Analyze at least 30 worms per strain to count the seam cells, and three independent experiments are performed.
- Count the total numbers of hypodermal seam cells per worm and calculate the average, SEM, and P-value with two-tailed Student’s t-test using GraphPad Prism.
Notes
- The genotype of JR672 is wIs54 [Pscm::gfp] V. Note that Pscm::gfp transgene is located on chromosome V.
- The GFP signal is stronger in the larval stage than in adults. Thus, select the GFP-expressing worm as soon as they reached the L4 stage.
Recipes
- Nematode Growth Media (NGM) Agar (2 L)
- Add the following ingredients in a 4 L flask:
NaCl 6.0 g
Agar, Bacteriological, Ultrapure 34 g
Bacto peptone 5 g
dH2O 1,950 ml
Note: Worms do not grow well on plates prepared with agar of lower grade. - Add a stir bar and mix well. Leave the stir bar inside of the flask
- Prepare 1 L of dH2O to wash the tubes for PourBoy® 4 Sterile Media Dispenser, and wrap the tubes with aluminum foil
- Autoclave NGM, dH2O and the tubes for PourBoy® 4 Sterile Media Dispenser using liquid 45 min cycle in Amsco® Century SV-120 Scientific Prevacuum Sterilizer
- Cool down the NGM on the stir plate until it reaches approximately 55 °C. Meanwhile, prepare 200 of 60 mm Petri-dishes
- Once NGM cools down, add the following ingredients:
2 ml of 1 M CaCl2
4 ml of 0.5 M MgSO4
50 ml of Potassium Phosphate (pH 6) and 2 ml Cholesterol (5 mg/ml in ethanol) - Mix well by stirring. It is okay that NGM turns cloudy
- Dispense 10 ml of NGM into 60 mm Petri-dish using PourBoy® 4 Sterile Media Dispenser. The NGM plates are dried for a couple of days at RT
- Seed OP50-1 as a food source for C. elegans. Add approximately 100 μl of OP50-1 in each plate and spread the bacteria using a home-made cell spreader to make a small circle of bacterial lawn. The bacterial lawn should not touch the edge of the plate. Then, culture the OP50-1 overnight at RT
- Add the following ingredients in a 4 L flask:
- 1 M CaCl2
- Dissolve 21.9 g of Calcium chloride hexahydrate (CaCl2•6H2O) in 90 ml dH2O
- Once it is dissolved, add more dH2O to make 100 ml of 1 M CaCl2
- Sterilize it by filtering using 0.22 μm filter units
- 0.5 M MgSO4
- Dissolve 12.03 g of Magnesium sulfate (MgSO4) in 90 ml dH2O
- Once it is dissolved, add more dH2O to make 100 ml of 0.5 M MgSO4
- Autoclave it with a liquid 45 min cycle in Amsco® Century SV-120 Scientific Prevacuum Sterilizer
- 1 M Potassium phosphate (pH 6)
- Prepare 100 ml of 1 M K2HPO4 by dissolving 17.42 g of potassium phosphate dibasic in dH2O
- Prepare 100 ml of 1 M KH2PO4 by dissolving 13.61 g of potassium phosphate monobasic in dH2O
- Take 13.2 ml of 1 M K2HPO4 and add 86.8 ml of 1 M KH2PO4 to make 1 M Potassium phosphate
- Autoclave 1 M potassium phosphate with a liquid 45 min cycle in Amsco® Century SV-120 Scientific Prevacuum Sterilizer
- 5 mg/ml cholesterol
Dissolve 0.5 g of cholesterol in 100 ml of ethanol - LB agar
- Add the following ingredients:
NaCl 2.5 g
Tryptone 2.5 g
Yeast Extract 1.25 g
Agar 3.75 g
dH2O up to 250 ml - Mix well with a stir bar and autoclave with a liquid 45 min cycle
- Cool it down to 55 °C and add 250 μl of 50 mg/ml streptomycin
- Stir it on the stir plate and pour into 100 mm Petri-dish
- Dry the plates on the bench overnight
- Add the following ingredients:
- LB
- Add the following ingredients:
NaCl 10 g
Tryptone 10 g
Yeast Extract 5 g
dH2O up to 1 L - Dissolve by stirring and autoclave with a liquid 45 min cycle
- Add the following ingredients:
- OP50-1 culture
- Streak OP50-1 on the LB agar plate containing streptomycin
- Culture OP50-1 in 37 °C incubator overnight
- Pick one colony of OP50-1 from the LB agar plate and culture in 20 ml of LB containing streptomycin at the final concentration of 50 μg/ml at 37 °C shaking incubator overnight
Acknowledgments
This work was supported by grants from NIH (NS074324, NS089616), The Robert Packard Center for ALS Research at Johns Hopkins, The ALS Association, and The Muscular Dystrophy Association.
Competing interests
The authors have no conflicts of interest or competing interests to declare.
References
- Altun, Z. F. and Hall, D. H. (2009). Epithelial system, hypodermis. In: WormAtlas. Doi:10.3098/wormatlas.1.13.
- Ambros, V. (1989). A hierarchy of regulatory genes controls a larva-to-adult developmental switch in C. elegans. Cell 57(1): 49-57.
- Bartel, D. P. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281-297.
- Bernstein, E., Caudy, A. A., Hammond, S. M. and Hannon, G. J. (2001). Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409(6818): 363-366.
- Bracht, J., Hunter, S., Eachus, R., Weeks, P. and Pasquinelli, A. E. (2004). Trans-splicing and polyadenylation of let-7 microRNA primary transcripts. RNA 10(10): 1586-1594.
- Carmell, M. A., Xuan, Z., Zhang, M. Q. and Hannon, G. J. (2002). The Argonaute family: tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes Dev 16(21): 2733-2742.
- Caudy, A. A., Myers, M., Hannon, G. J. and Hammond, S. M. (2002). Fragile X-related protein and VIG associate with the RNA interference machinery. Genes Dev 16(19): 2491-2496.
- Caudy, A. A., Ketting, R. F., Hammond, S. M., Denli, A. M., Bathoorn, A. M., Tops, B. B., Silva, J. M., Myers, M. M., Hannon, G. J. and Plasterk, R. H. (2003). A micrococcal nuclease homologue in RNAi effector complexes. Nature 425(6956): 411-414.
- Fielenbach, N. and Antebi, A. (2008). C. elegans dauer formation and the molecular basis of plasticity. Genes Dev 22(16): 2149-2165.
- Grishok, A., Pasquinelli, A. E., Conte, D., Li, N., Parrish, S., Ha, I., Baillie, D. L., Fire, A., Ruvkun, G. and Mello, C. C. (2001). Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106(1): 23-34.
- Hammond, S. M., Boettcher, S., Caudy, A. A., Kobayashi, R. and Hannon, G. J. (2001). Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 293(5532): 1146-1150.
- Hong, Y., Lee, R. C. and Ambros, V. (2000). Structure and function analysis of LIN-14, a temporal regulator of postembryonic developmental events in Caenorhabditis elegans. Mol Cell Biol 20(6): 2285-2295.
- Huynh, J. M., Dang, H. and Fares, H. (2018). Measurement of lysosomal size and lysosomal marker intensities in adult Caenorhabditis elegans. Bio-protocol 8 (3): e2724.
- Koh, K. and Rothman, J. H. (2001). ELT-5 and ELT-6 are required continuously to regulate epidermal seam cell differentiation and cell fusion in C. elegans. Development 128:2867-2880.
- Lee, R. C., Feinbaum, R. L. and Ambros, V. (1993). The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75(5): 843-854.
- Lee, Y., Jeon, K., Lee, J. T., Kim, S. and Kim, V. N. (2002). MicroRNA maturation: stepwise processing and subcellular localization. EMBO J 21(17): 4663-4670.
- Lee, Y., Kim, M., Han, J., Yeom, K. H., Lee, S., Baek, S. H. and Kim, V. N. (2004). MicroRNA genes are transcribed by RNA polymerase II. EMBO J 23(20): 4051-4060.
- Mourelatos, Z., Dostie, J., Paushkin, S., Sharma, A., Charroux, B., Abel, L., Rappsilber, J., Mann, M. and Dreyfuss, G. (2002). miRNPs: a novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev 16(6): 720-728.
- Reinhart, B. J., Slack, F. J., Basson, M., Pasquinelli, A. E., Bettinger, J. C., Rougvie, A. E., Horvitz, H. R. and Ruvkun, G. (2000). The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403(6772): 901-906.
- Ruvkun, G. and Giusto, J. (1989). The Caenorhabditis elegans heterochronic gene lin-14 encodes a nuclear protein that forms a temporal developmental switch. Nature 338(6213): 313-319.
- Singh, R. N. and Sulston, J. E. (1978). Some observations on moulting in Caenorhabditis elegans. Nematologica 24: 63-71.
- Slack, F. J., Basson, M., Liu, Z., Ambros, V., Horvitz, H. R. and Ruvkun, G. (2000). The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol Cell 5(4): 659-669.
- Terns, R. M., Kroll-Conner, P., Zhu, J., Chung, S and Rothman, J. H. (1997). A seficiency screen for zygotic Loci required for establishment and patterning of the epidermis in Caenorhabditis elegans. Genetics 146(1):185-206.
- Thein, M. C., McCormack, G., Winter, A. D., Johnstone, I. L., Shoemaker, C. B. and Page, A. P. (2003). Caenorhabditis elegans exoskeleton collagen COL-19: an adult-specific marker for collagen modification and assembly, and the analysis of organismal morphology. Dev Dyn 226(3): 523-539.
- Yi, R., Qin, Y., Macara, I. G. and Cullen, B. R. (2003). Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev 17(24): 3011-3016.
- Zhang, T., Wu, Y. C., Mullane, P., Ji, Y. J., Liu, H., He, L., Arora, A., Hwang, H. Y., Alessi, A. F., Niaki, A. G., Periz, G., Guo, L., Wang, H., Elkayam, E., Joshua-Tor, L., Myong, S., Kim, J. K., Shorter, J., Ong, S. E., Leung, A. K. L. and Wang, J. (2018). FUS regulates activity of microRNA-mediated gene silencing. Mol Cell 69(5): 787-801 e788.
Article Information
Copyright
© 2019 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Ji, Y. J. and Wang, J. (2019). Heterochronic Phenotype Analysis of Hypodermal Seam Cells in Caenorhabditis elegans. Bio-protocol 9(1): e3132. DOI: 10.21769/BioProtoc.3132.
Category
Developmental Biology > Cell growth and fate > Differentiation
Neuroscience > Cellular mechanisms > Neuronal fate
Cell Biology > Cell-based analysis > Heterochronic phenotype
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link