- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Measurement of Lysosomal Size and Lysosomal Marker Intensities in Adult Caenorhabditis elegans
Published: Vol 8, Iss 3, Feb 5, 2018 DOI: 10.21769/BioProtoc.2724 Views: 7334
Reviewed by: Manish ChamoliAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Simultaneous Monitoring Cytoplasmic Calcium Ion and Cell Surface Phosphatidylserine in the Necrotic Touch Neurons of Caenorhabditis elegans
Yoshitaka Furuta and Zheng Zhou
Oct 20, 2021 2624 Views

Live-cell Imaging and Analysis of Germline Stem Cell Mitosis in Caenorhabditis elegans
Réda M. Zellag [...] Abigail R. Gerhold
Jan 5, 2022 4696 Views

SunTag-Based Single-Molecule Translation Imaging in Caenorhabditis elegans
Elise van der Salm [...] Suzan Ruijtenberg
Oct 20, 2025 2284 Views
Abstract
Assays have been developed to study trafficking in various tissues of Caenorhabditis elegans. Adult C. elegans intestinal cells are large and have extensive endocytic networks, thus making them a good system for deciphering the endocytic pathway using live imaging techniques. However, the presence of auto-fluorescent gut granules in adult intestine can interfere with the signals of endocytic compartment reporters, like GFP. Here we demonstrate a protocol adapted from the original method developed by the Grant laboratory to identify signals from reporters in adult intestinal cells. The goal of this protocol is to identify endocytic compartments tagged with fluorescent markers without any confounding effects of background autofluorescent gut granules in adult intestinal cells of Caenorhabditis elegans.
Keywords: C. elegansBackground
Caenorhabditis elegans is a multicellular organism that has been used to study endocytic trafficking. Originally, assays were developed to study endocytosis in C. elegans oocytes, embryos, and coelomocytes (scavenger cells). Briefly, the assays in oocytes and embryos were performed by measuring the intensities and sizes of compartments containing a yolk protein-green fluorescent protein reporter (VIT-2::GFP) in intestinal compartments at the comma to ‘1.5 fold’ stages of development (Grant and Hirsh, 1999; Schaheen et al., 2006a). In adults, the intensities and sizes of compartments containing GFP (secreted from body wall muscle cells into the psuedocoelom and endocytosed by coelomocytes) were measured in the coelomocytes of transgenic adult C. elegans expressing Pmyo-3::ssGFP (signal sequence-GFP fusion protein) (Treusch et al., 2004). These assays have been used to identify and to elucidate functions of mediators of the endocytic pathway (Fares and Greenwald, 2001a and 2001b; Schaheen et al., 2006b; Huynh et al., 2016).
Here, we present an assay that can be used to study endocytosis in another C. elegans tissue. Adult intestinal cells of C. elegans are large and are thus also a great system for deciphering the endocytic pathway using live imaging techniques. The functions of intestinal cells include food assimilation and synthesis, storage of macromolecules, stress response, and host-pathogen interactions (McGhee, 2007). However, one of the main challenges of studying endocytic transport by live imaging in adult intestinal cells is the prevalence of auto-fluorescent gut granules that interfere with the unambiguous determination of bona fide endocytic compartment reporter (like GFP) signals and therefore bias qualitative and quantitative studies (Clokey and Jacobson, 1986). We therefore adapted a method developed by the Grant laboratory to conclusively identify signals from reporters in adult intestinal cells (Gleason et al., 2016; Huynh et al., 2016).
Materials and Reagents
- Microscope slides (VWR, catalog number: 48382-171 )
- Coverslips for microscope slides (Fisher Scientific, Fisherbrand, catalog number: 12-541A )
- 60 mm plates (Fisher Scientific, Fisherbrand, catalog number: FB0875713A )
- 100 mm plates (Fisher Scientific, Fisherbrand, catalog number: FB0875713 )
- Labeling tape (Fisher Scientific, Fisherbrand, catalog number: 15-901-5K )
- Glass pipette
- Aluminum foil
- Autoclave tape
- Inoculating loops
- Pipette tips
- C. elegans experimental strain:
RT258: unc-119(ed3); pwIs50[lmp-1::GFP, unc-119]
Note: LMP-1 is the orthologue of mammalian Lamp1 that localizes to lysosomes in Caenorhabditis elegans (Kostich et al., 2000) - C. elegans control strain: N2
- Calcium chloride dihydrate (CaCl2·2H2O) (Fisher Scientific, catalog number: C79-500 )
- Magnesium sulfate heptahydrate (MgSO4·7H2O) (Fisher Scientific, catalog number: M63-500 )
- Potassium phosphate monobasic (KH2PO4) (Fisher Scientific, catalog number P285-500 )
- Cholesterol (Sigma-Aldrich, catalog number: C8667 )
- EtOH (Merck, catalog number: EX0276-4 )
- LE agarose for making 2.2% agarose (BioExpress, GeneMate, catalog number: E-3120 )
- Levamisole (Sigma-Aldrich, catalog number: 31742 )
- C. elegans culture
- Platinum wire-pick to transfer C. elegans
- Making NGM plates (Brenner, 1974; He, 2011) (see Recipes)
Sodium chloride (NaCl) (Fisher Scientific, catalog number: S271-3 )
Bacto peptone (BD, BactoTM, catalog number: 211677 )
Bacto agar (BD, BactoTM, catalog number: 214030 )
Double distilled water
Cholesterol 5 mg/ml in 95% EtOH (see Recipes)
1 M CaCl2 sterile (see Recipes)
1 M MgSO4 sterile (see Recipes)
1 M KH2PO4 pH 6.0 sterile (see Recipes) - Making 2x YT + OP50 for spotting NGM plates
OP50 frozen stock
2x YT agar plate - 2x YT agar plate (see Recipes)
Bacto tryptone (BD, BactoTM, catalog number: 211705 )
Yeast extract (Fisher Scientific, catalog number: BP1422-500 )
Sodium chloride (NaCl) (Fisher Scientific, catalog number: S271-3 )
Bacto agar (BD, catalog number: 214030 )
Double distilled water - 2x YT medium (see Recipes)
- 2x YT medium + OP50 (see Recipes)
- Platinum wire-pick to transfer C. elegans
- 2.2% agarose pad (see Recipes)
- 9 mM levamisole/1x PBS (see Recipes)
Equipment
- 20 °C Incubator for C. elegans storage (VWR, manufactured by Sheldon Manufacturing, model: Model 2020 )
- 4 L flask
- Stir bar
- Stir plate
- Autoclave
- 2 L beaker
- 1 L bottle
- 37 °C Incubator (VWR, manufactured by Sheldon Manufacturing, model: 5025 T )
- Microwave
- Heating block
- 5-100 ml bottle
- Microscope (Carl Zeiss, model: STEMI SV 6 )
- Zeiss LSM 510 Meta confocal microscope (Zeiss, model: LSM 510 ):
- 63x lens
- Argon 488-nm laser
- Helium neon 543-nm laser
- 63x lens
Software
- MetaMorph® Microscopy Automation & Image Analysis Software (Sunnyvale, CA)
Procedure
Note: Before starting the protocol: Ensure that 60 mm NGM plates (spotted with OP50) have been prepared.
Day 1
- Set up three plates with three adult hermaphrodites (one day post-L4 stage) on each spotted 60 mm NGM plate for each C. elegans experimental and control strain.
- Leave plates in a 20 °C incubator for three days for the hermaphrodites to lay eggs.
Day 4
Move 50-100 L4 stage C. elegans of each experimental strain onto new OP50-spotted 60 mm NGM plates (the offspring of the P0 hermaphrodites). This ensures that these animals would be fairly synchronized young adults for imaging on the following day.
Day 5: Preparation for microscopy
- Prepare the imaging slide by first making the agarose pad. See steps 1-4 in Figure 1 for instructions.
- Next, add 7 μl of 9 mM levamisole/1x PBS (see Recipes) to the center of the agarose pad (Figure 1, S5).
- Pick 20-30 adult worms of one C. elegans experimental strain and submerge them in the 9 mM levamisole/1x PBS (Figure 1, S5).
- Put a cover slip on top of the agarose pad, ensuring that it fully covers the C. elegans and the 9 mM levamisole/1x PBS (Figure 1, S6).
- Repeat Steps 1-5 for all experimental and control strains used.
- Do microscopy to image LMP-1::GFP (or marker of interest) and focus on the intestine right below the pharynx, using the 63x objective. Take images that show the desired signal while avoiding saturation of the signal.
- Image C. elegans wild type strain RT258 first with both the Argon 488-nm laser and the Helium neon 543-nm laser. RT258 should be imaged first because it is the wild type strain which measurements from other experimental strains should be compared against.
- Take images of the intestines of approximately 5-10 adult C. elegans for the wild type RT258 strain.
- Repeat Steps 7 and 8 for each control and experimental strain. Make sure that images of all strains are taken using the same exposure and magnification as RT258.

Figure 1. Preparation of imaging slides
Data analysis
- MetaMorph version 7.5.0.0 was used to analyze confocal microscopy images, though other software could also be used.
- All confocal microscopy images were converted to ‘.tif’ format for use with MetaMorph.
- Identify the compartments that ONLY have green fluorescence (excited by the 488-nm laser). These compartments are LMP-1::GFP-positive that marks lysosomes. Compartments that are yellow (fluoresce with both 488-nm and 543-nm lasers) are gut granules, which will also be seen in C. elegans control strain N2: these gut granule compartments should not be measured or included for determining sizes of bona fide LMP-1::GFP compartments (Figure 2). We do not know whether the LMP-1::GFP signal decreases in older adults; if it does, this protocol may not be effective at differentiating bona fide GFP signal from autofluorescence.
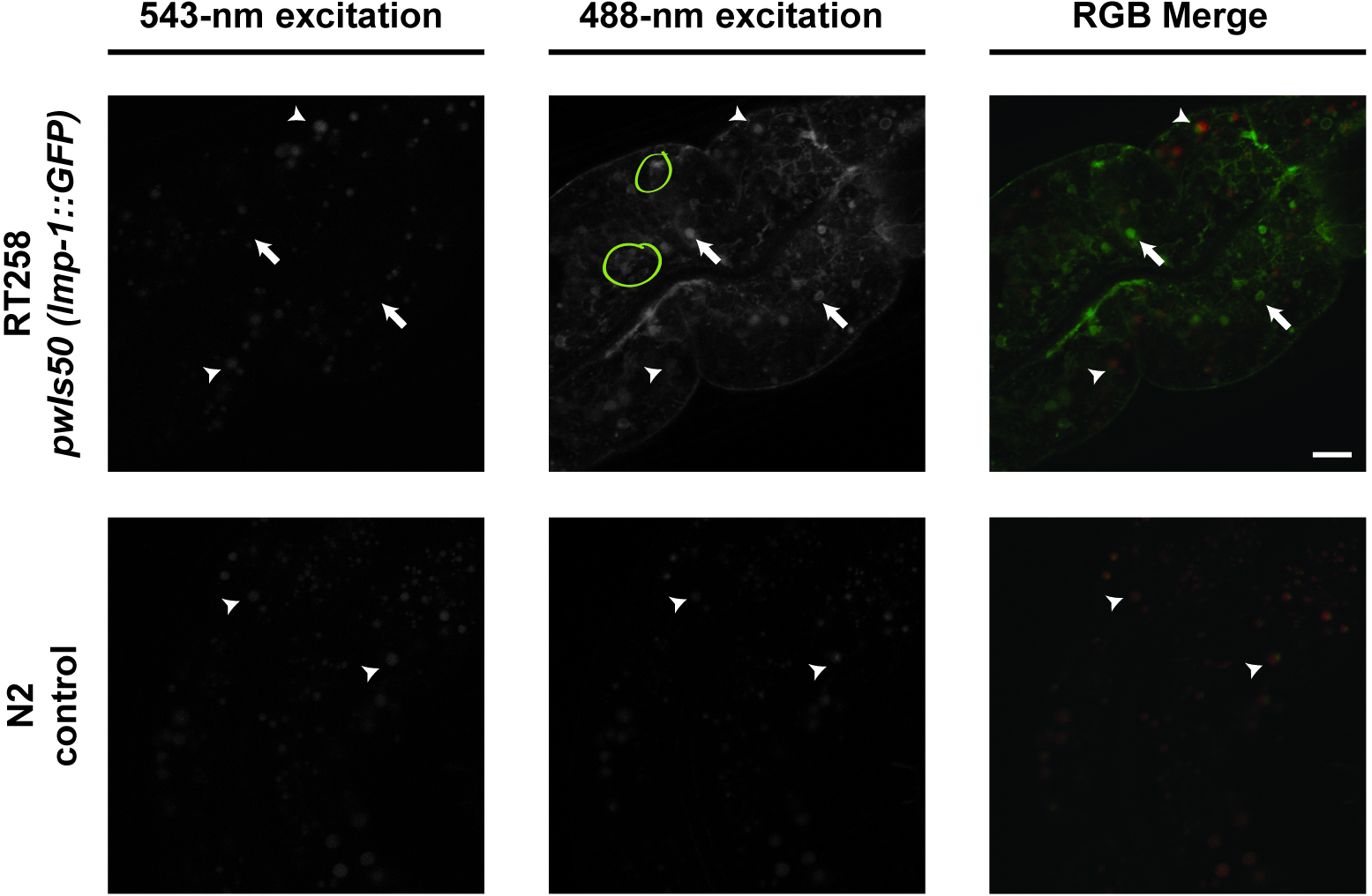
Figure 2. Representative images of intestinal images. Compartments that fluoresce with both 543-nm and 488-nm excitation are granules while compartments that fluoresce with only 488-nm excitation are LMP-1::GFP-positive compartments. Arrows indicate examples of LMP-1::GFP-positive compartments. Arrowheads indicate examples of gut granules. Yellow circles indicate clumped compartments. Scale bar = 10 μm. - For each LMP-1::GFP positive compartment, use MetaMorph to measure the intensity and area of the selected compartment. See Figure 3 for a pictorial guide (Figure 3).
- Go to Measure in the tool bar and select ‘Show Region Statistics…’
- Use the trace region tool to select the LMP-1::GFP positive compartment.
- Double click on the selected region.
- To get measurements of LMP-1::GFP-positive compartments’ intensities, use the ‘Average’ Gray Level value. This value is already normalized by the area size.
- To get measurements of LMP-1::GFP-positive compartment size, use the ‘Area’ value.
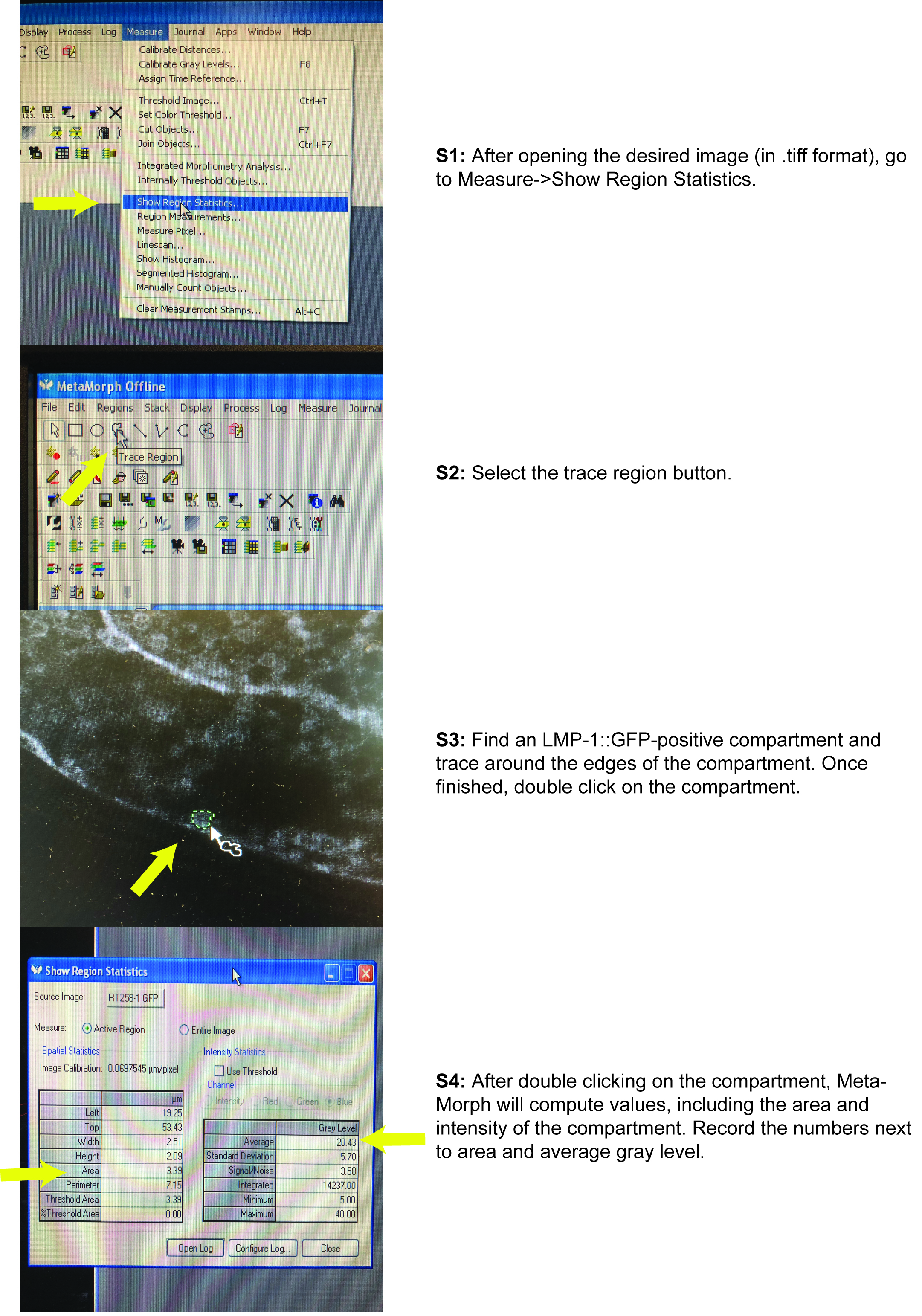
Figure 3. Using MetaMorph to measure LMP-1::GFP-positive compartment size and marker intensity - Collect measurements of all the LMP-1::GFP-positive compartments for each adult C. elegans intestine, for each strain, from each image. Do not include clumped compartments because it is difficult to get accurate quantitations (due to ambiguity in the location of each individual compartment). The result is approximately 10-20 clearly defined compartments per image.
- Average all the LMP-1::GFP-positive intensity measurements using Excel to obtain the average LMP-1::GFP-positive intensity for each strain. Average all of the LMP-1::GFP positive size measurements to obtain the average compartment size for each strain. Figure 4 depicts results obtained when comparing the wild type strain with a cup-5(null) strain (CUP-5 is a cation channel; loss of CUP-5 protein results in lysosomal dysfunction) (Fares and Greenwald, 2001b; Hersh et al., 2002). Loss of CUP-5 resulted in an increase in LMP-1::GFP-positive intensity, but no significant change in lysosomal size.
- Go to Measure in the tool bar and select ‘Show Region Statistics…’
Note: We have successfully used this protocol on strains carrying various mutations or following RNAi treatment.
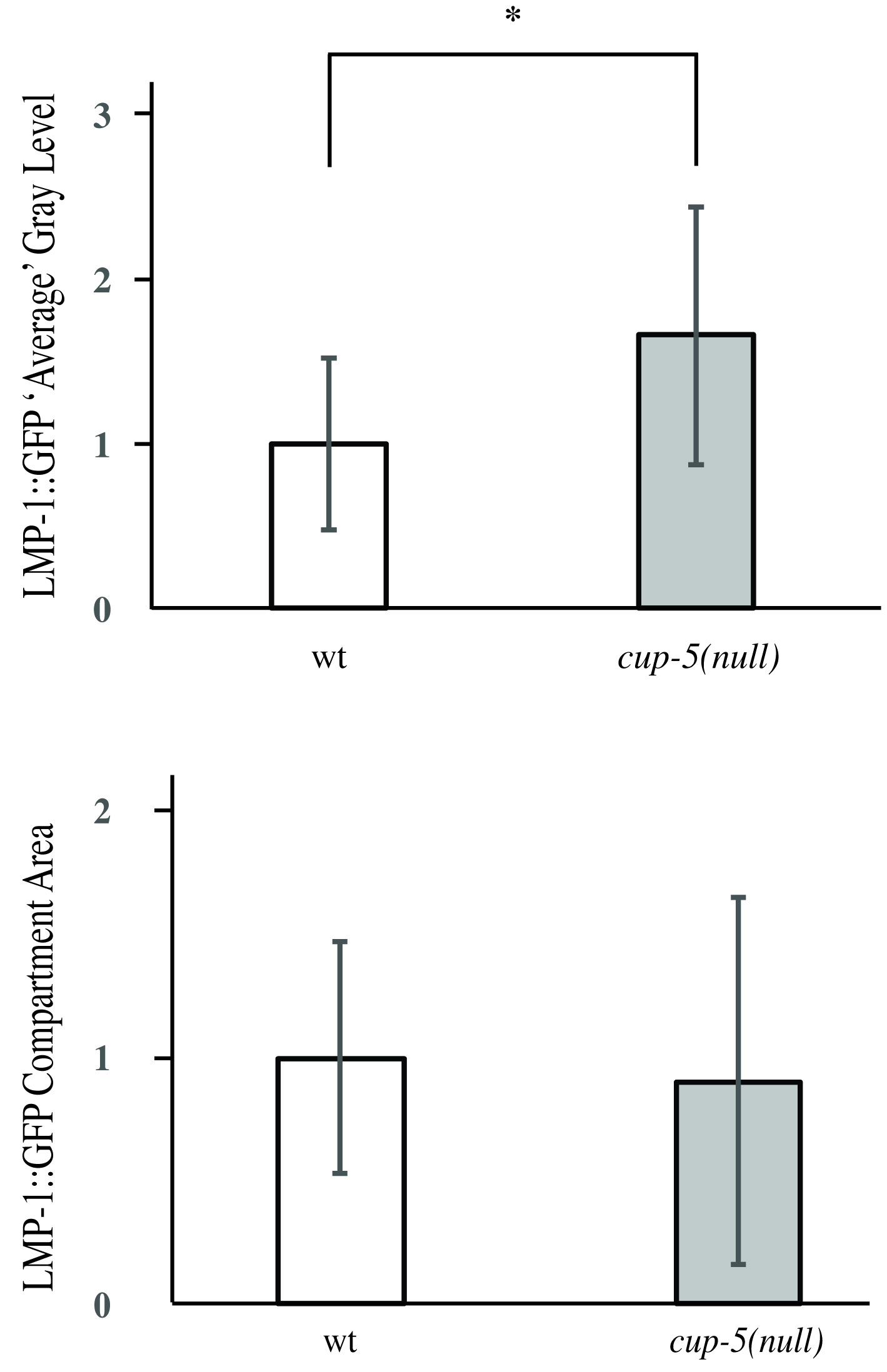
Figure 4. Representative images of LMP-1::GFP intensity and compartment size data obtained. * indicates P < 0.05.
Notes
- The higher the ‘Average’ Gray Level, the larger the lysosomal compartment size.
- If imaging multiple strains with the same marker, take all images using identical confocal settings, preferably during the same microscopy session. This would allow comparisons of compartment sizes and intensities between strains/genotypes. Additionally, though not always technically possible, when feasible, strains to be compared should be imaged on the same slide to reduce slide-to-slide variation during microscopy.
- Repeat this experiment multiple times to determine reproducibility and statistical significance.
- This method can be used to analyze markers for other organelles than lysosomes in adult intestinal cells, distinguishing marker signals from gut granule autofluorescence.
Recipes
- Nematode Growth Medium (NGM) agar (3 L)
- Add the following to a 4-L flask:
NaCl (9.0 g)
Bacto peptone (7.5 g)
Bacto agar (51 g)
Double distilled water (2,925 ml) - Stir bar
- Autoclave for 1 h
- Place flask on stir plate, set on low stir, and let cool to 55 °C
- Once cool, add the following:
CaCl2 1 M sterile (3 ml)
MgSO4 1 M sterile (3 ml)
KH2PO4 1 M pH 6.0 sterile (75 ml)
Cholesterol 5 mg/ml in 95% EtOH (3 ml) - Pour 8 ml into each 60 mm plate
- Let dry for one night before storing unspotted plates at 4 °C or spotting with 2x YT + OP50
- Add the following to a 4-L flask:
- 2x YT agar (1 L)
- Add the following into a 2-L (or larger) flask:
NaCl (5.0 g)
Bacto tryptone (16 g)
Yeast extract (10 g)
Bacto agar (15.0 g)
Double distilled water (Up to 1 L) - Stir bar
- Autoclave for 1 h
- Place flask on stir plate, set on low stir, and let cool 55 °C
- Pour 18 ml into each 100 mm plate
- Wait two days for plates to dry and then store at 4 °C
- Add the following into a 2-L (or larger) flask:
- 2x YT medium (1 L)
- Add the following to a 2-L beaker
Tryptone (16 g)
Yeast extract (10 g)
NaCl (5 g)
Fill to 1 L with double distilled water - Add a stir bar into the beaker and let the mixture stir on a stir plate until homogenous
- Pour into 1 L bottles
- Autoclave for 30 min
- Store at room temperature
- Add the following to a 2-L beaker
- 2x YT medium + OP50 (1 L)
- Day 1: Use a pipette tip to spread OP50 frozen stock across a 2x YT agar plate. Place plate in a 37 °C incubator and leave it overnight
- Day 2: Use an inoculating loop to pick one colony from the 2x YT agar plate and transplant the colony to a 1 L bottle of 2x YT Medium. Place the 1 L bottle into the 37 °C incubator and leave it overnight
- Day 3: Remove bottle from the 37 °C incubator and store at 4 °C until use
- Day 1: Use a pipette tip to spread OP50 frozen stock across a 2x YT agar plate. Place plate in a 37 °C incubator and leave it overnight
- 2.2% agarose pad
- Add the following to a flask:
Agarose (0.22 g)
Double distilled water (10 ml) - Microwave flask for 60 sec. If the agarose and double distilled water solution is not homogenous, microwave for another 30 sec
- Store in a tube that is in a heating block (68 °C)
- Add the following to a flask:
- 9 mM levamisole/1x PBS
- Mix the following in a tube:
9 ml of 10 mM levamisole (Sigma-Aldrich)
1 ml of 10x PBS - Store at room temperature
- Mix the following in a tube:
- Cholesterol (5 mg/ml)
- Add the following to a 500 ml beaker
Cholesterol (2.5 g)
100% EtOH (475 ml)
Double distilled water (25 ml) - Add a stir bar in the beaker and let the mixture stir on a stir plate until homogenous
- Pour 100 ml into five 100 ml bottle (please confirm this description)
- Add the following to a 500 ml beaker
- 1 M CaCl2
- In a beaker, dissolve CaCl2 (73.5 g) into 500 ml double distilled water
- Add a Stir bar into the beaker and let the mixture stir on a stir plate until homogenous
- Pour into a 500 ml bottle
- Autoclave for 30 min
- In a beaker, dissolve CaCl2 (73.5 g) into 500 ml double distilled water
- 1 M MgSO4
- In a beaker, dissolve MgSO4 (123.25 g) into 500 ml double distilled water
- Add a Stir bar into the beaker and let the mixture stir on a stir plate until homogenous
- Pour into a 500 ml bottle
- Autoclave for 30 min
- In a beaker, dissolve MgSO4 (123.25 g) into 500 ml double distilled water
- 1 M KH2PO4 pH 6.0
- In a beaker, dissolve KH2PO4 (68.05 g) into 450 ml with double distilled water
- Add a Stir bar into the beaker and let the mixture stir on a stir plate until homogenous
- Adjust pH to 6.0 with NaOH
- Fill up to 500 ml with double distilled water
- Pour into a 500 ml bottle
- Autoclave for 30 min
- In a beaker, dissolve KH2PO4 (68.05 g) into 450 ml with double distilled water
Acknowledgments
This protocol was adapted from Huynh et al. (2016). This work was supported by a Microscopy Society of America grant (to J.M.H) and by National Science Foundation grant 3004290 (to H.F.). The authors have no conflicts of interest or competing interests.
References
- Brenner, S. (1974). The genetics of Caenorhabditis elegans. Genetics 77(1): 71-94.
- Clokey, G. V. and Jacobson, L. A. (1986). The autofluorescent “lipofuscin granules” in the intestinal cells of Caenorhabditis elegans are secondary lysosomes. Mech Ageing Dev 35(1): 79-94.
- Fares, H. and Greenwald, I. (2001a). Genetic analysis of endocytosis in Caenorhabditis elegans: coelomocyte uptake defective mutants. Genetics 159(1): 133-145.
- Fares, H. and Greenwald, I. (2001b). Regulation of endocytosis by CUP-5, the Caenorhabditis elegans mucolipin-1 homolog. Nat Genet 28(1): 64-68.
- Gleason, A. M., Nguyen, K. C., Hall, D. H. and Grant, B. D. (2016). Syndapin/SDPN-1 is required for endocytic recycling and endosomal actin association in the C. elegans intestine. Mol Biol Cell.
- Grant, B. and Hirsh, D. (1999). Receptor-mediated endocytosis in the Caenorhabditis elegans oocyte. Mol Biol Cell 10(12): 4311-4326.
- He, F. (2011). Common worm media and buffers. Bio-protocol Bio101: e55.
- Hersh, B. M., Hartwieg, E. and Horvitz, H. R. (2002). The Caenorhabditis elegans mucolipin-like gene cup-5 is essential for viability and regulates lysosomes in multiple cell types. Proc Natl Acad Sci U S A 99(7): 4355-4360.
- Huynh, J. M., Dang, H., Munoz-Tucker, I. A., O'Ketch, M., Liu, I. T., Perno, S., Bhuyan, N., Crain, A., Borbon, I. and Fares, H. (2016). ESCRT-dependent cell death in a Caenorhabditis elegans model of the lysosomal storage disorder mucolipidosis type IV. Genetics 202(2): 619-638.
- Kostich, M., Fire, A. and Fambrough, D. M. (2000). Identification and molecular-genetic characterization of a LAMP/CD68-like protein from Caenorhabditis elegans. J Cell Sci 113 ( Pt 14): 2595-2606.
- McGhee, J. D. (2007). The C. elegans intestine. WormBook: 1-36.
- Schaheen, L., Dang, H. and Fares, H. (2006a). Basis of lethality in C. elegans lacking CUP-5, the Mucolipidosis Type IV orthologue. Dev Biol 293(2): 382-391.
- Schaheen, L., Patton, G. and Fares, H. (2006b). Suppression of the cup-5 mucolipidosis type IV-related lysosomal dysfunction by the inactivation of an ABC transporter in C. elegans. Development 133(19): 3939-3948.
- Treusch, S., Knuth, S., Slaugenhaupt, S. A., Goldin, E., Grant, B. D. and Fares, H. (2004). Caenorhabditis elegans functional orthologue of human protein h-mucolipin-1 is required for lysosome biogenesis. Proc Natl Acad Sci U S A 101(13): 4483-4488.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Huynh, J. M., Dang, H. and Fares, H. (2018). Measurement of Lysosomal Size and Lysosomal Marker Intensities in Adult Caenorhabditis elegans. Bio-protocol 8(3): e2724. DOI: 10.21769/BioProtoc.2724.
Category
Cell Biology > Cell imaging > Live-cell imaging
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link