Advanced Search
STAG1-AID , STAG2-AID
Last updated date: May 8, 2020 Views: 1161 Forks: 0
Design gRNAs using online tools: https://zlab.bio/guide-design-resources
EGFP-AID-STAG1 gRNA1: CACCGTCTTCAGACTTCAGAACAT, gRNA2: CACCGGTTTCTCATCATTTTTCTA.
EGFP-AID-STAG2 gRNA1: CACCGCACAGATTTAATTGTGTAC, gRNA2: CACCGCTCTCTCTCATTAGGTTCT.
Oligo annealing and cloning into gRNA expression plasmid PX335:
1. Digest 1µg of PX335 with BbsI for 30 min at 37°C:
1 µg PX335
1 µl FastDigest BbsI (Fermentas)
1 µl FastAP (Fermentas)
2 µl 10X FastDigest Buffer
X µl ddH2O
20 µl total
2. Gel purify digested plasmid using QIAquick Gel Extraction Kit and elute in EB.
3. Phosphorylate and anneal each pair of oligos:
1 µl oligo 1 (100uM)
1 µl oligo 2 (100uM)
1 µl 10X T4 Ligation Buffer (NEB)
6.5 µl ddH2O
0.5 µl T4 PNK (NEB)
10 µl total
Anneal in a thermocycler using the following parameters:
37°C 30 min, 95°C 5 min and then ramp down to 25°C at 5°C/min
4. Set up ligation reaction and incubate at room temperature for 10 min:
X µl BbsI digested PX335 (50ng)
1 µl phosphorylated and annealed oligo duplex (1:200 dilution)
5 µl 2X Quickligation Buffer (NEB)
X µl ddH2O
10 µl subtotal
1 ul Quick Ligase (NEB)
11 µl total
5. Transformation to E.coli
6. Validate the sequence of gRNA expression plasmid by Sanger sequencing using the primer 5’-
ACCTCGACCATGGTAATAGCG
template DNA cloning
1. PCR N-terminal TSS containing region of STAG1/STAG2 by Phusion Hot Start II DNA Polymerase:
10 µl 5X Phusion HF Buffer
1 µl 10 mM dNTPs
1 µl Primer pair (10 uM)
0.5 µl HeLa genomic DNA (20 ng/ul)
0.5 µl Phusion Hot Start
37 µl ddH2O
50 µl total
98 °C 30s
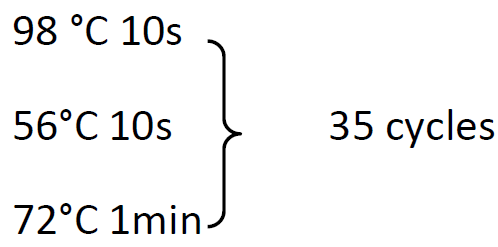
72°C 5min
2. Gel purified PCR product ligated to pJET vector
5 µl 2X Reaction Buffer
2 µl purified PCR product (20 ng/µL)
0.5 µl pJET1.2/blunt Cloning Vector (50 ng/µL)
0.5 µl T4 DNA Ligase
2 µl ddH2O
10 µl total
Incubate the ligation mixture at room temperature (22°C) for 10 min.
3. transformation to E. coli
4. Validate the sequence of gRNA expression plasmid by Sanger sequencing using the primer fw: 5'-CGACTCACTATAGGGAGAGCGGC-3' and rev: 5'-AAGAACATCGATTTTCCATGGCAG-3’
Transfection of HeLa cells
1. 1 day prior to transfection, seed HeLa cells in 10 cm2 dish containing 10 mL of complete growth medium without Penicillin/Streptomycin. Grow cells overnight to approximately 50%-60% confluence.
2. Mix 4 μg template DNA, 2 μg of each gRNA with 1 mL Opti-MEM® I.
3. Mix 40 μl Lipofectamine 2000 with 1 mL Opti-MEM® I. Incubate the mixture for 5 minutes at room temperature.
4. Combine the diluted DNA with the diluted transfection reagent. Incubate at room temperature for 20 min.
5. Add the DNA-transfection reagent complexes to the cell and mix gently.
6. Incubate the cells for 24h, split the cells with fresh medium containing Penicillin/Streptomycin.
7. GFP positive single cell is sorted
8. A week later singe cell colony could be used for genotyping.
Identification of homozygous knock-in cells by genotyping
1. Genomic DNA isolation by nexttec™ 1-Step DNA Isolation Systems: https://www.nexttec.de/products/tissue-and-cells
- Add 140 µl Buffer G and 10 µl Proteinase K to each sample. Incubate the sample with shaking (56°C, 200 rpm, 30 min to overnight)
- Transfer 100 µl of the lysate to an equilibrated nexttec™ cleanPlate96. Incubate for 3 min at room temperature. Centrifuge at 700x g for 1 min or apply vacuum for 1 min.
2. PCR amplification of the CRISPR targeted region
4 µl 5X Phusion HF Buffer
0.4 µl 10 mM dNTPs
0.4 µl Primer pair (10 uM)
1 µl HeLa genomic DNA (20 ng/ul)
0.2 µl Phusion Hot Start
14 µl ddH2O
20 µl total
98 °C 30s
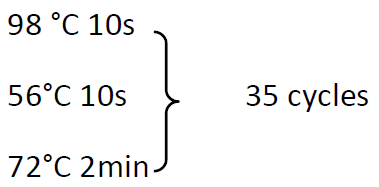
72°C 5min
EGFP-AID-STAG1 primers used for genotyping were: forward primer:GCCAGCTGGGAATCTCTTCA, and reverse primer:GCCACAGTTTGCTGACTCCT.
EGFP-AID-STAG2 primers used for genotyping were: forward primer:AGAAAGAAGGCAAGCCACCA and reverse primer: GGCAGCAGGAAGTACCTAACT.
- Tang, W(2020). STAG1-AID , STAG2-AID. Bio-protocol Preprint. bio-protocol.org/prep307.
- Wutz, G., Ladurner, R., St Hilaire, B. G., Stocsits, R. R., Nagasaka, K., Pignard, B., Sanborn, A., Tang, W., Várnai, C., Ivanov, M. P., Schoenfelder, S., van der Lelij, P., Huang, X., Dürnberger, G., Roitinger, E., Mechtler, K., Davidson, I. F., Fraser, P., Lieberman-Aiden, E. and Peters, J.(2020). ESCO1 and CTCF enable formation of long chromatin loops by protecting cohesinSTAG1 from WAPL. eLife. DOI: 10.7554/eLife.52091
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link
