- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
A Streamlined Method for the Preparation of Gelatin Embedded Brains and Simplified Organization of Sections for Serial Reconstructions
Published: Vol 7, Iss 22, Nov 20, 2017 DOI: 10.21769/BioProtoc.2610 Views: 14387
Reviewed by: Shai BerlinJunjie LuoYunbing Ma

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Single-Particle Tracking of AMPA Receptor-Containing Vesicles
Victor C. Wong [...] Erin K. O’Shea
Jun 5, 2025 2260 Views

Local Iontophoretic Application for Pharmacological Induction of Long-Term Synaptic Depression
Borys Olifirov [...] Pavel Belan
Jun 5, 2025 1897 Views

Ultrafast Isolation of Synaptic Terminals From Rat Brain for Cryo-Electron Tomography Analysis
Rong Sun and Qiangjun Zhou
Sep 5, 2025 3556 Views
Abstract
Gelatin embedding of whole brains for sectioning is a critical procedure used in neuroscience to ensure all morphological and spatial details are preserved intact. Here, we describe an inexpensive, reproducible and efficient means to embed post-fixed brains ready for sectioning in gelatin within a week’s time. The sections obtained are distortion-free and their fragile internal structures preserved which can be used for serial reconstructions for lesion studies and mapping of viral expression after stereotaxic injections. In addition, the separation of adjacent slices into a series of 3-4 vials facilitates subsequent organization and assembly of serial sections at the mounting step.
Keywords: Gelatin embeddingBackground
Recent advances in behavioral neurosciences have allowed the introduction of opsins and antibody targeted-toxins to specific subsets of neurons and regions of the brain. These studies often require visualization of whole brain sections for histological and morphological analysis to localize cell-type specific antibody targeted-toxin induced lesions by stereotaxic injection for behavioral validation (Aoki et al., 2015) or the introduction of virus delivered transgenes (Aquili et al., 2014). Detailed mapping of neuronal circuitry by rabies virus (Suzuki et al., 2012) and localization surveys of serial sections have been achieved using gelatin as an embedding agent, which acts a structural substrate within and around the tissue. Gelatin impregnated brain tissue provides strengthened support for delicate internal structures such as the hippocampus and ventricle spaces which are easily damaged when processed for immunohistochemistry (IHC) and subsequent mounting onto slides. Embedded brain sections are also often free of distortions when mounted and adjacent sections can be used for serial reconstructions.
Qualities of the gel embedding and post-IHC handling are dependent on several procedural details, which can be tedious and time consuming. In previous published protocols, proper penetration and infiltration of the gelatin into the brain and ventricular spaces required a vacuum oven (Griffioen et al., 1992). Further, embedding the brains in a small mold is challenging as the brain has a tendency to float. Additionally, identifying and orienting adjacent slices after IHC processing for serial reconstructions can be arduous.
In this improved protocol, we address several issues important for timely and trouble-free gelatin embedding of whole brains. Begin with rapid impregnation of gelatin into the brain using a magnetic stir bar to weigh the brain down into the liquefied gelatin. Further, the use of an icebox allows the gelatin to set from bottom to top thus eliminating the problem of floating brains. Finally, the arrangement and determination of adjacent serial sections for 3D reconstruction series are simplified by placing the adjacent sections sequentially into 3 to 4 vials, looping back to the first vial after the last. After IHC processing, the sections from each vial are placed in a column next to one another in a large Petri-dish and adjacent slices can be rapidly mounted moving along row by row.
Materials and Reagents
- 50 ml conical tubes in styrofoam frack (Corning, Falcon®, catalog number: 352098 )
- Weigh boats, small and medium (Dyn-A-Med Plus, catalog numbers: 80051 , 80056 )
- 150 mm Petri dishes (Corning, Falcon®, catalog number: 351058 )
- PS-10 vials (AS ONE, catalog number: 9-892-12 )
- Frosted slides (Matsunami Glass, catalog number: S024410 )
- Single edge razors (FEATHER Safety Razor, catalog number: 99129 )
- Paintbrush (Arteje Brush Camlon Pro, model 630 #3/0 Round)
- Kimwipe
- Paraformaldehyde (Merck, catalog number: 104005 )
- Sodium phosphate dibasic (Na2HPO4) (Wako Pure Chemical Industries, catalog number: 197-09705 )
- Sodium phosphate monobasic (NaH2PO4) (Wako Pure Chemical Industries, catalog number: 197-02865 )
- Sucrose (Wako Pure Chemical Industries, catalog number: 190-00013 )
- Gelatin (Wako Pure Chemical Industries, catalog number: 077-03155 )
- H2O2 (Wako Pure Chemical Industries, catalog number: 081-04215 )
- Triton X-100 (Bio-Rad Laboratories, catalog number: 1610407 )
- Tween-20 (Bio-Rad Laboratories, catalog number: 1706531 )
- Anti-tyrosine hydroxylase (Enzo Life Sciences, catalog number: BML-SA497-0100 )
- Thionin (Alfa Aesar, catalog number: A18912 )
- Standard ABC Peroxidase Kit (Vector Laboratories, catalog number: PK-4000 )
- Metal Enhanced DAB Kit (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 34065 )
- Entellan new mounting medium (Merck, catalog number: 107961 )
- Rabbit Anti-ChAT conjugated saporin (Advanced Targeting Systems, catalog number: IT-42 )
- Mouse Anti-NeuN (Abcam, catalog number: ab104224 )
- Goat Anti-ChAT (EMD Millipore, catalog number: AB144P )
- Biotin conjugated Goat anti-rabbit IgG (Thermo Fisher Scientific, catalog number: B-2770 )
- Biotin conjugated Goat anti-mouse IgG (Thermo Fisher Scientific, catalog number: B-2763 )
- Biotin conjugated Rabbit anti-goat IgG (Thermo Fisher Scientific, catalog number: A10518 )
- 4% PFA/0.1 M PB (4% paraformaldehyde/0.1 M phosphate buffer, pH 7.4) (see Recipes)
- 0.1 M PB (0.1 M phosphate buffer, pH 7.4) (see Recipes)
- 30% sucrose 0.1 M phosphate buffer, pH 7.4 (see Recipes)
- 4% paraformaldehyde/10% sucrose in 0.1 M phosphate buffer, pH 7.4 (see Recipes)
- 0.05 M PB (0.05 M phosphate buffer, pH 7.4) (see Recipes)
- 30% sucrose 0.05 M phosphate buffer, pH 7.4 (see Recipes)
- Blocking solution (see Recipes)
- Antibody solution (see Recipes)
- 0.5% w/v gelatin in H2O (see Recipes)
Equipment
- Rack for 50 ml conical tubes
- Thermometer
- Spoon spatula
- Spin-plus 19 mm magnetic stir-bars (SP Scienceware - Bel-Art Products - H-B Instrument, catalog number: F37144-0034 ) and magnetic spin-bar removal tool
- 2 x 500 ml beakers (DWK Life Sciences, Duran®, catalog number: 21 106 48 )
- 5 L liquid waste buckets for PFA/gel (AS ONE, catalog number: 4-5308-03 )
- Bent forceps (Ideal-Tek, model: 650.S.6 )
- Oven (SANYO, model number: MIR-162 )
- Hot plate with magnetic stirring capability (IKA, model: C-MAG HS 7 )
- Refrigerator (SANYO, model: MPR-414FR )
- Fume hood
- Vibratome (Leica Biosystems, model: Leica VT1000 S )
- Upright light microscope (Olympus, model: CX22LED )
Procedure
- Gelatin embedding
- Wash 4% PFA/0.1 M PB (see Recipes), perfused, 72 h post-fixed brains, once with 0.1 M phosphate buffer (PB) (see Recipes) and immerse brain in 30% sucrose/0.1 M PB (see Recipes) in a 50 ml conical tube. Use magnetic stir-bar atop a 15 ml inverted conical tube cap to help keep brain submerged in the tube. Incubate at 4 °C overnight (18 h).
- Decant 30% sucrose/0.1 M PB and replace with 50 °C double distilled water (ddH2O) and put tubes containing brains in a 55 °C incubator for 45 min to equilibrate the temperature of the brains. Continue to use magnetic stir-bar and conical cap to submerge brain for all subsequent steps between 50-55 °C.
- While brains are being temperature equilibrated, pre-warm hot plate for gelatin preparation, make sure temperature of the hot plate does not exceed 60 °C.
- Make 14% gelatin/10% sucrose solution (25 ml/brain); for 500 ml (20 brains), add 70 g gelatin little by little to 450 ml of 50 °C ddH2O using magnetic stir bar speed from slow to fast, which will pull in gelatin without causing clumps (Video 1).
- Once the gelatin is dissolved, add 50 g of sucrose and bring volume to 500 ml with 50 °C ddH2O.
- Make 7% gelatin/5% sucrose solution by pre-warming 150 ml of ddH2O to 55 °C and add 150 ml of 14% gelatin/10% sucrose to the pre-warmed ddH2O, keep all gelatin solutions at 55 °C in oven (Video 1).Video 1. Gelatin solution preparation. Visual demonstration of 14% gelatin/10% sucrose and 7% gelatin/5% sucrose preparation is shown in this example.
- Decant warm ddH2O submerging the brains in 7% gelatin solution/5% sucrose, swirl, incubate at 55 °C for 2 h (Video 2).Video 2. Brain submersion in gelatin. Visual demonstration of removal of solutions and submersion in gelatin using magnetic stir bar is shown in this example.
- Decant 7% gelatin solution/5% sucrose submerging the brains in 14% gelatin solution/10% sucrose, swirl, and incubate at 55 °C for 3 h.
- Mark weigh boats with animal ID numbers.
- Using a spoon spatula, work swiftly and carefully dislodge brain and liquid contents into a weigh boat atop a large Petri dish on a bed of ice. Top off with gelatin, remove bubbles, orient the brain and gently press down with bent forceps to keep it submerged until the bottom begins to set (about 7-10 min, Video 3).Video 3. Casting of gelatin embedded brain and removal after gelatin setting. Visual demonstration of casting gelatin embedded brain in weigh boat mold and subsequent cutting of set mold from weigh boat are shown in this example.
- Move brains to a 4 °C refrigerator and leave for another 90 min.
- Mark sample vials with animal ID numbers.
- Carefully cut out the casted brain with a new single edge razor blade and slide into sample vial.
- Add 4% PFA/10% sucrose in 0.1 M PB (see Recipes) to the vial with brain, place the vial in a refrigerator overnight (18 h).
- Remove PFA, rinse once with 0.05 M PB (see Recipes) and replace with 30% sucrose/0.05 M PB (see Recipes). Keep sample vials in the refrigerator for a minimum of 72 h.
- Wash 4% PFA/0.1 M PB (see Recipes), perfused, 72 h post-fixed brains, once with 0.1 M phosphate buffer (PB) (see Recipes) and immerse brain in 30% sucrose/0.1 M PB (see Recipes) in a 50 ml conical tube. Use magnetic stir-bar atop a 15 ml inverted conical tube cap to help keep brain submerged in the tube. Incubate at 4 °C overnight (18 h).
- Sequential slice collection
- After embedding, mark 3 to 4 vials for each animal. (i.e., animal ID–#1 to 3 or 4)
- Cut block that includes the region of interest, trim block and systematically cut diagonal corners to orient the slices correctly during mounting (i.e., upper right and lower left corners relative to the face of the block).
- Collect sections from microtome or vibratome in 0.05 M PB and place sections sequentially among the 3 to 4 vials. Loop back to the first vial after the third or fourth vial (Figure 1). Sections between 40-80 μm from the vibratome generally work the best.

Figure 1. Slice collection schematic from sectioning device and post-immunostaining arrangement of slices at the mounting step. Slices are collected sequentially in numbered vials as shown in the figure. After the completion of immunostaining procedure, each of the vial’s slices is arranged in columns in a large Petri dish, with the next vial’s column arranged beside the previous vial. Once arranged, serial mounting is achieved by moving along row by row. Arrangement by sequential vial collection is simpler since the distance between slices is greater. This allows for slices to be easily distinguished from one another more accurately and thus, mounting is performed more rapidly.
- After embedding, mark 3 to 4 vials for each animal. (i.e., animal ID–#1 to 3 or 4)
- Immunostaining
Users’ choice: fluorescent labeling or IHC. Briefly, the examples shown in Figures 2 and 3 were stained by the following procedures.- Pretreat the samples with 3% H2O2 in 0.05 M PB for 20 min, and then incubate the samples with blocking solution (see Recipes) for 1 h.
- Decant the blocking solution and add primaries in antibody solution (see Recipes, 1:100 [anti-ChAT]; 1:1,000 [anti-TH], 1:1,000 [anti-NeuN]), incubate at 4 °C for 48 h.
- Decant the primaries. Wash with PBS for 5 min, 3 times.
- Add biotin-conjugated secondary antibodies in antibody solution (1:400 anti-goat, ChAT, anti-rabbit for TH and anti-mouse for NeuN), incubate at 25 °C for 4 h.
- Decant the secondaries. Wash with PBS for 5 min, 3 times.
- Add Standard ABC Peroxidase solution diluted according to manufacturer’s protocol in PBS and 0.2% Tween-20, incubate at 25 °C for 4 h.
- Decant the ABC Peroxidase solution. Wash with PBS for 5 min, 3 times.
- Make DAB working solution according to the manufacturer’s protocol and add to slices. Wait approximately 3-5 min for signal development and stop reaction with four 0.1 M PB washes.
- Decant ABC peroxidase solution and wash with PBS for 5 min, 3 times.
- Store slices in 0.05 M PB.
- Pretreat the samples with 3% H2O2 in 0.05 M PB for 20 min, and then incubate the samples with blocking solution (see Recipes) for 1 h.
- Serial mounting after immunostaining
- Dump contents of first vial into large Petri dish with 0.01 M PB (1:5 dilution of 0.05 M PB in H2O) and arrange in a column using a fine paintbrush.
- Carefully move contents of second vial and arrange in column next to first vial’s column.
- Repeat until all vials are arranged in columns next to one another.
- Mount adjacent serial slices on slides with a paintbrush, moving along row by row (Figures 1 and 2).
- If dry mounting IHC slices, prepare slide by coating with 0.12% w/v gelatin in ddH2O (dilute 1:4 0.5% w/v gelatin in ddH2O, see Recipes) with a paintbrush in a medium sized weigh boat. Leave slide half submerged by resting the frosted side on the square edge of the weigh boat and the submerged side on the bottom slope of the spout (Video 4).
- Move initial slices from large Petri dish to submerged slide in medium-sized weigh boat containing 0.12% gelatin by placing slice in the solution, orienting the slice and then carefully sliding the slice up to its proper position away from the submerged edge of the slide.
- Push the slide towards the spout of the weigh boat, exposing more of the slide from the 0.12% gelatin and repeating the process in step D6 for the adjacent slices to fill the rest of the slide.
- Blot dry edge of slide on Kimwipe and air dry for 24 h at 22 °C before counterstaining, dehydration, mounting medium and cover slip placement.
Video 4. Mounting of sections on a slide. Visual demonstration of mounting gelatin embedded brain slices in a weigh boat filled with 0.12% w/v gelatin. In this example, only one of four vials was used for mapping antibody-mediated toxin deletions (each slice is 240 μm apart) and sequentially arranged from top left, moving along top to bottom, left to right.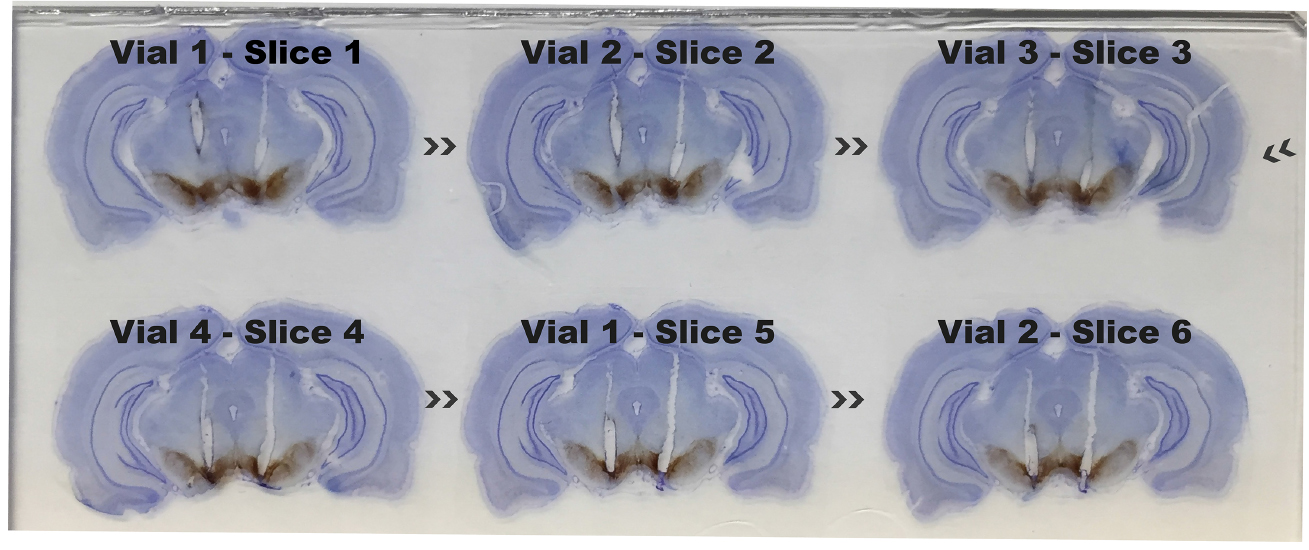
Figure 2. Cannula placement localized in the ventral tegmental area (VTA) using serial sections. Gelatin embedded rat brain slices were stained with anti-tyrosine hydroxylase visualized with horseradish peroxidase and 3’,3’-diaminobenzidine (DAB). Slices were dry mounted onto a slide, counterstained with 0.02% thionin for 3 minutes, run through an ethanol dehydration series and cover-slipped with Entellan mounting medium. Six 80 μm serial slices were mounted sequentially starting from top left moving along horizontally ending on the bottom right and the vial and slice number are labeled above the slice (see Figure 1 for origin of vial and slice).
- Dump contents of first vial into large Petri dish with 0.01 M PB (1:5 dilution of 0.05 M PB in H2O) and arrange in a column using a fine paintbrush.
Data analysis
Localization of cannula placement in the rat VTA was verified using an Olympus CX22LED light microscope (Figure 2). Additionally, verification and mapping of the extent of stereotaxically injected anti-ChAT-saporin antibody directed lesions in the rat striatum (Aoki et al., 2015, Figure 3) were analyzed qualitatively by using an Olympus CX22LED light microscope and anatomical landmarks from a rat brain atlas (Paxinos and Watson, 2004).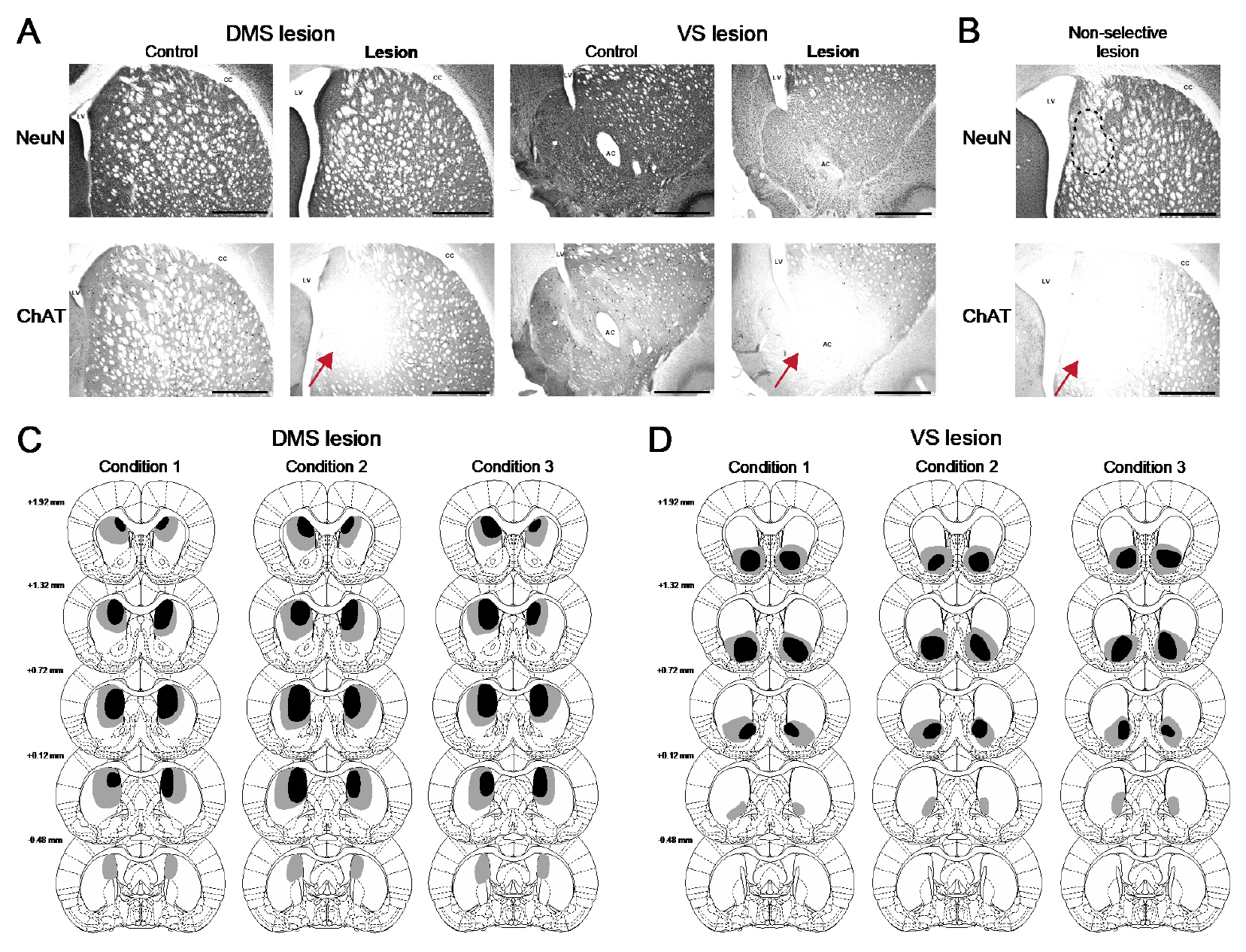
Figure 3. Verification and lesion mapping of antibody targeted immunotoxin delivery to cholinergic neurons in the rat striatum. A. Representative coronal sections of rat striatal DAB stained slices display mostly intact NeuN staining but cell-specific deletion of cholinergic interneurons in ChAT staining among rabbit anti-ChAT-saporin mediated lesioned cases (arrows–dorsal medial stratum, DMS or ventral striatum, VS). Scale bars = 1 mm. B. An example of a limited nonspecific lesion indicated by a dashed area (NeuN) where fewer cells appear and an example more extensive non-specific deletion using a non-selective goat IgG-saporin injection as a control (arrow). C and D. The most restricted (solid black) and the broadest areas (gray) of cholinergic interneuronal deletions in each of the behavioral conditions are shown. Coordinate distances of the slices from the bregma are indicated to the left. The extent of the lesions in DMS (C) and VS (D) appears similarly between all conditions. Anatomical landmarks in the DAB stained photographs: LV, lateral ventricle; CC, corpus callosum; AC, anterior commissure. Figure modified for clarification and reprinted with permission (Aoki et al., 2015).
Notes
It is strongly suggested that vials and weigh boats are marked before hand with animal IDs and that the transfer of the brain from one container to another is done carefully as to ensure there is no confusion of the identity of the individual.
Recipes
- 4% PFA/0.1 M PB (4% paraformaldehyde/0.1 M phosphate buffer, pH 7.4)
4% w/v paraformaldehyde in ddH2O
80 mM Na2HPO4
20 mM NaH2PO4 - 0.1 M PB (0.1 M phosphate buffer, pH 7.4)
80 mM Na2HPO4 in ddH2O
20 mM NaH2PO4 - 30% sucrose/0.1 M PB (30% sucrose 0.1 M phosphate buffer, pH 7.4)
30% w/v sucrose in ddH2O
80 mM Na2HPO4
20 mM NaH2PO4 - 4% paraformaldehyde/10% sucrose in 0.1 M phosphate buffer, pH 7.4
4% w/v paraformaldehyde in ddH2O
20% w/v sucrose
80 mM Na2HPO4
20 mM NaH2PO4 - 0.05 M PB (0.05 M phosphate buffer, pH 7.4)
40 mM Na2HPO4 in ddH2O
10 mM NaH2PO4 - 30% sucrose 0.05 M phosphate buffer, pH 7.4
30% w/v sucrose in ddH2O
40 mM Na2HPO4
10 mM NaH2PO4 - Blocking solution
1x PBS
5% secondary’s host serum
0.2% Triton X-100 - Antibody solution
1x PBS
2% secondary’s host serum
0.2% Triton X-100 - 0.5% w/v gelatin in ddH2O
ddH2O to 50 °C
0.5% w/v gelatin
Acknowledgments
We thank Dr. Tom Ruigrok who provided the initial protocols for gelatin embedding and Mayank Aggarwal for allowing us to display the stained slide of cannula placement in the VTA used in Figure 2. We also greatly appreciate François Beauchain for critical proofreading and Yumiko Akamine for assistance with the video production. Finally, we tip our hats to Andres Carrasco for calling our attention Bio-protocol.
Support from the Human Frontier Science Program (A.W.L., J.R.W.), JSPS Grant-in-Aid for Challenging Exploratory Research, Grant-in-Aid for JSPS Fellows and Grant-in-Aid for Young Scientists–category A (S.A.) made research performed in this publication possible. The authors declare no competing financial interests.
References
- Aoki, S., Liu, A. W., Zucca, A., Zucca, S. and Wickens, J. R. (2015). Role of striatal cholinergic interneurons in set-shifting in the rat. J Neurosci 35(25): 9424-9431.
- Aquili, L., Liu, A. W., Shindou, M., Shindou, T. and Wickens, J. R. (2014). Behavioral flexibility is increased by optogenetic inhibition of neurons in the nucleus accumbens shell during specific time segments. Learn Mem 21(4): 223-231.
- Griffioen, H. A., Van der Beek, E. and Boer, G. J. (1992). Gelatin embedding to preserve lesion-damaged hypothalami and intracerebroventricular grafts for vibratome slicing and immunocytochemistry. J Neurosci Methods 43(1): 43-47.
- Paxinos, G. and Watson, C. (2004). The rat brain in stereotaxic coordinates. Elsevier Academic Press.
- Suzuki, L., Coulon, P., Sabel-Goedknegt, E. H. and Ruigrok, T. J. (2012). Organization of cerebral projections to identified cerebellar zones in the posterior cerebellum of the rat. J Neurosci 32(32): 10854-10869.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Liu, A. W., Aoki, S. and Wickens, J. R. (2017). A Streamlined Method for the Preparation of Gelatin Embedded Brains and Simplified Organization of Sections for Serial Reconstructions. Bio-protocol 7(22): e2610. DOI: 10.21769/BioProtoc.2610.
- Aoki, S., Liu, A. W., Zucca, A., Zucca, S. and Wickens, J. R. (2015). Role of striatal cholinergic interneurons in set-shifting in the rat. J Neurosci 35(25): 9424-9431.
Category
Neuroscience > Cellular mechanisms > Synaptic physiology
Neuroscience > Neuroanatomy and circuitry > Animal model
Cell Biology > Cell imaging > Fixed-tissue imaging
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link