- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Histochemical Preparations to Depict the Structure of Cauliflower Leaf Hydathodes
Published: Vol 7, Iss 20, Oct 20, 2017 DOI: 10.21769/BioProtoc.2452 Views: 8530
Reviewed by: Hiroyuki HiraiYunbing MaAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Live-Cell Monitoring of Piecemeal Chloroplast Autophagy
Masanori Izumi [...] Shinya Hagihara
Nov 5, 2025 1688 Views

Chloroplast Movement Imaging Under Different Light Regimes With a Hyperspectral Camera
Paweł Hermanowicz [...] Justyna Łabuz
Dec 20, 2025 749 Views

Reproducible Emu-Based Workflow for High-Fidelity Soil and Plant Microbiome Profiling on HPC Clusters
Henrique M. Dias [...] Christopher Graham
Jan 20, 2026 403 Views
Abstract
Hydathodes are plant organs present on leaf margins of a wide range of vascular plants and are the sites of guttation. Both anatomy and physiology of hydathodes are poorly documented. We have recently reported on the anatomy of cauliflower and Arabidopsis thaliana hydathodes and on their infection by the vascular pathogenic bacterium Xanthomonas campestris pv. campestris (Xcc) (Cerutti et al., 2017). Because hydathodes are natural infection routes for several pathogens, it is necessary to have a deep knowledge of their anatomy to further better interpret images of infected hydathodes. Here, we described different detailed protocols for gaining information on hydathode anatomy which are applicable to a wide range of plants (including monocots like barley and rice). Nomarsky and confocal microscopy were used to observe clarified thick samples. Optical microscopy in transmitted light and transmission electron microscopy were used to observed thin and ultrathin sections.
Keywords: CauliflowerBackground
In literature, different techniques were used to study hydathodes (Perrin, 1972; Chen and Chen, 2007; Wang et al., 2011; Singh, 2014). From light microscopy (on entire tissues or on section of resin-embedded samples) to scanning or transmission electron microscopy, a large panel of protocols and techniques was available. To our knowledge, these techniques were not used in combination and laser confocal microscopy was never used to depict hydathode structures. Moreover, we noticed variations from protocols to protocols. We presented here different techniques used in combination. They are well-adapted to cauliflower and Arabidopsis thaliana. They have been successfully applied to other plants like monocotyledons (barley and rice) and should be likely used to a larger variety of plant species. We encourage the users to apply these protocols to gain complementary information on the hydathode at different scales during infection.
Materials and Reagents
- Microscope glass slides (Thermo Fisher Scientific, Superfrost, catalog number: 10143560WCUT )
- Cover slip 24 x 60 mm (Thermo Fisher Scientific, catalog number: 15747592 )
- Razor blades
- Cauliflower (Brassica oleracea var. botrytis, cultivar Clovis)
Notes:- Cauliflower plants were grown in a controlled greenhouse.
- All the experiments used the second true leaf from four-weeks-old plants.
- Cauliflower plants were grown in a controlled greenhouse.
- Chloral hydrate (Sigma-Aldrich, catalog number: 23100 )
- Glycerol (C3H8O3) (VWR, catalog number: 24387.326 )
- Calcofluor (Fluorescent Brightener 28) (Sigma-Aldrich, catalog number: F3543 )
- Triton X-100 (Sigma-Aldrich, catalog number: T8787 )
- Sodium cacodylate (Sigma-Aldrich, catalog number: C4945 )
- Glutaraldehyde EM grade (Electron Microscopy Sciences, catalog number: 16214 )
- Osmium tetroxide (OsO4) (Electron Microscopy Sciences, catalog number: 19150 )
- Ethanol (C2H5OH) (Sigma-Aldrich, catalog number: 32221 )
- Epon (Electron Microscopy Sciences, catalog number: 14120 )
Note: Composition of the resin: Embed-812 (45 g), DDSA (36 g), NMA (18 g) and BDMA (1.35 ml). Embedding kit in which accelerator DMP30 is replaced by BDMA (Electron Microscopy Sciences, catalog number: 11400 ). - Borax (Sigma-Aldrich, catalog number: B3545 )
- Toluidine blue (RAL Diagnostics, catalog number: 361590 )
- Methylene blue (Merck, catalog number: 159270 )
- Basic fuchsin (Sigma-Aldrich, catalog number: 857343 )
- Periodic acid (VWR, Prolabo, catalog number: 20.593.151 )
- Acetic acid (CH3COOH) (CARLO ERBA Reagents, catalog number: 302016 )
- Thiocarbohydrazide (Sigma-Aldrich, catalog number: 223220 )
- Silver proteinate (Roques for histology) (Sigma-Aldrich, catalog number: 05495 )
- Aceton (CH3COCH3) (Fisher Scientific, catalog number: 10395640 )
- Clarification solution (see Recipes)
- Fixation solution (see Recipes)
- Sodium cacodylate buffer (see Recipes)
- Borax solution with toluidine blue and methylene blue (see Recipes)
- Basic fuchsin solution (see Recipes)
- Periodic acid solution (see Recipes)
- Silver proteinate solution (see Recipes)
Equipment
- Hollow punch (Harris, Uni-Core) (Electron Microscopy Sciences, catalog number: 69039-70 )
- Ultra-microtome (Leica Microsystems, Reichert-Jung, model: UltraCut E )
- Optical microscope equipped for Nomarski (Leica Microsystems, model: DM IRB-E )
- Confocal microscope (Leica Microsystems, model: Leica TCS SP2 ) equipped with a diode laser at 405 nm
- Diaphragm pump vacuum or other vacuum sources
- Stirrer hotplate (IKA)
- Microwave apparatus (Leica Microsystems, model: EM AMW )
- Flat bottom embedding capsule (Electron Microscopy Sciences, catalog number: 70021 )
- Oven (Memmert, model: UFE500BO )
- Histo diamond knife (Diatome Histo) for semi-thin sections (0.5 µm) to optical observations
- Ultra diamond knife (Diatome Ultra 45) for ultra-thin sections (70-80 nm) to transmission electron microscopy
- Hot plate (slides warmer) (C & A Scientific, Premiere, model: XH-2002 )
- Optical microscope (ZEISS, model: Axioplan 2 )
- CCD camera (ZEISS, model: AxioCam MRc )
- Transmission electron microscope (Hitachi, model: HT7700 )
- Gold grid (Electron Microscopy Sciences, catalog number: FCF200-Au )
Software
- Pro Plus 4.0 Imaging software (Media Cybernetics, Silver Spring, MD, USA)
Procedure
Nomarski and confocal microscopy were used to observe clarified thick samples to depict the complex vascularization and the epithem, respectively. Optical microscopy in transmitted light (on thin sections) and transmission electron microscopy (on ultrathin sections) were used to localize both bacteria within the hydathode and changes in cell or tissue structure during infection.
- Clarification of the samples for Nomarski and laser confocal microscopy
- Use the hollow punch or razor blades to take pieces of leaf margins.
- Immerse the samples in the chloral hydrate-based clarification solution (see Recipe 1) for several weeks (usually 1-3 weeks) at 4 °C in the dark (in a fridge).
- Mount samples in the same solution and observe by Nomarski microscopy. Acquire images (see Figure 1).
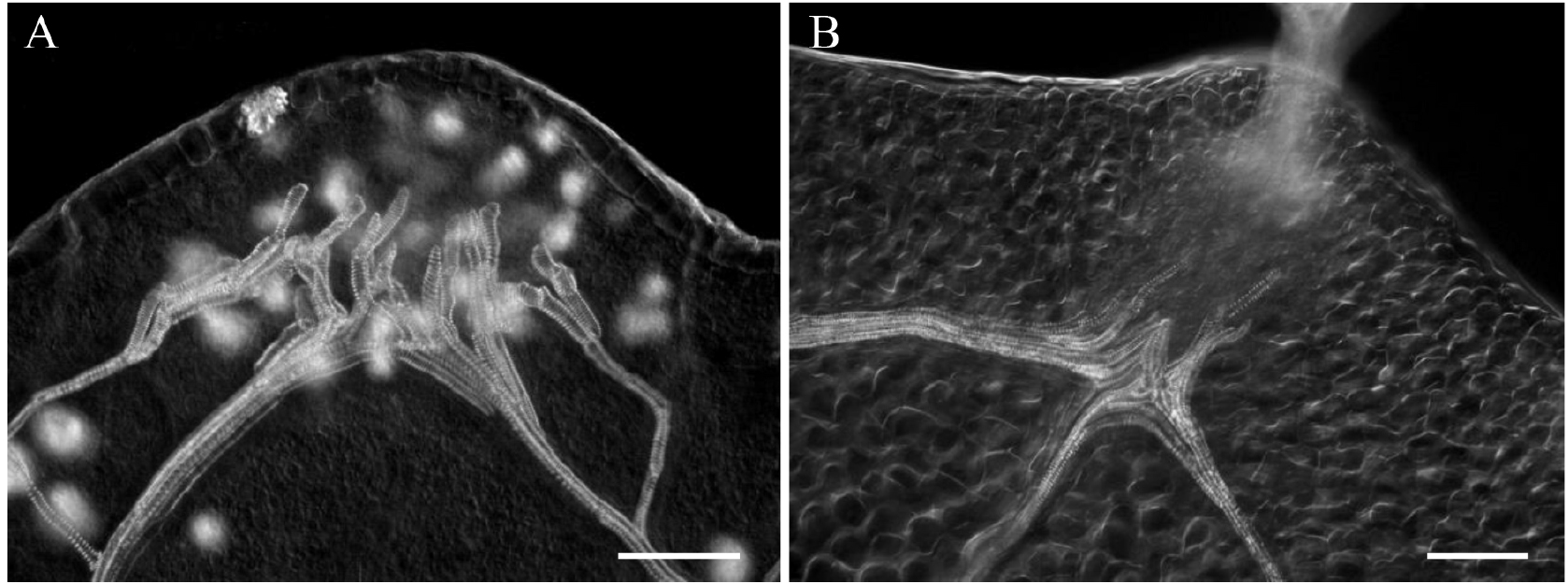
Figure 1. Typical images of cauliflower (A) and Arabidopsis (B) acquired in Nomarsky. Scale bars = 100 µm. - For confocal microscopy of the same samples, rinse the samples overnight in distilled water at 4 °C.
- Stain the samples in an aqueous solution of 0.01% calcofluor (for cellulose) for 1 h at room temperature.
- Rinse in distilled water and mount in water on a glass slide.
- Acquire images using the 405 nm ray line of laser diode for excitation and collect the emitted fluorescence between 410 and 500 nm. The samples being thick, we use objective lens with long working distance to avoid too much pressure on the sample during observation and acquisition. Thus, we favor water immersion objective lenses (working distance of 2 to 3 mm) to perform observations in depth (see Figure 2).
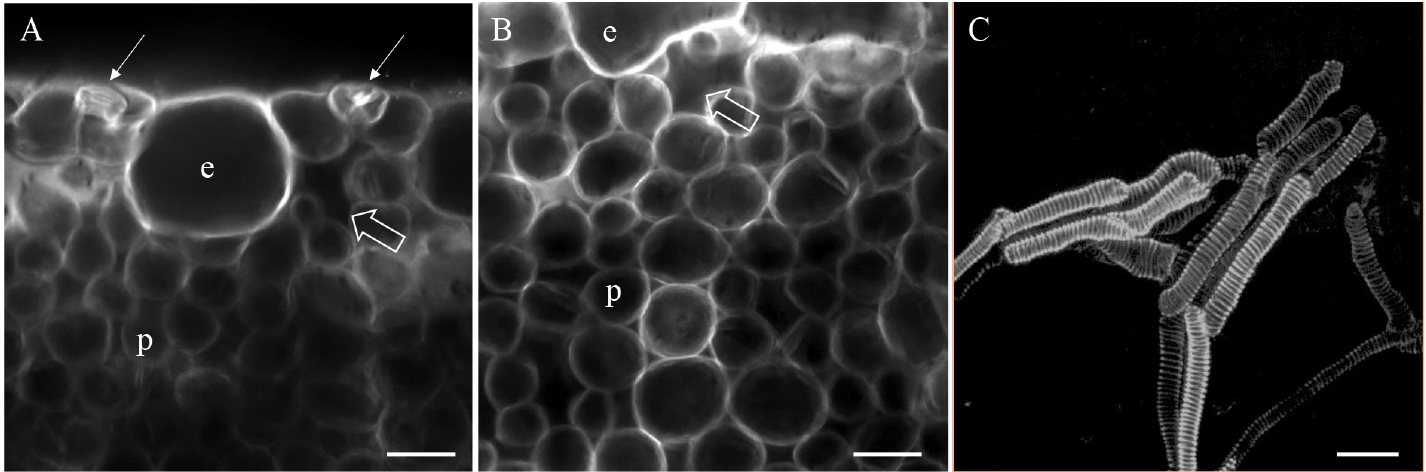
Figure 2. Confocal plane (A-B) of paradermal optical sectioning of cauliflower hydathode. A. View of the edge of the hydathode with two pores (arrows) and a large chamber below the pore (open arrow). B. Image of the epithem tissue of the hydathode with numerous meatuses. e: epidermal cell; p: parenchyma cell. C. Maximal projection of 20 confocal planes (z-stack) within a hydathode to depict the organization of the tracheids and their ornamentation. Scale bars = 25 µm.
- Use the hollow punch or razor blades to take pieces of leaf margins.
- Sample preparation for electron and optical microscopy
- With a razor blade, take pieces (2-3 mm2) of leaf margins encompassing hydathodes.
- Fix the samples under vacuum for 10 min with fixation solution (see Recipe 2). Release the vacuum slowly and repeat (2-3 times) until the samples sink.
- Fix the samples for 1 h, at the atmospheric pressure, in the fixation solution without Triton X-100.
- Rinse the samples three times in 0.2 M sodium cacodylate buffer (15 min each time, see Recipe 3).
- Post-fix the samples with 2% osmium tetroxide in 0.2 M sodium cacodylate buffer for 1 h at room temperature.
- Rinse the samples three times in 0.2 M sodium cacodylate buffer (15 min each time).
- Dehydrate the samples in a graded series of aqueous solution of ethanol concentrations (25%, 50%, 1 h for each treatment).
- Conserve the samples in ethanol 70% (minimum 1 h) until the use of Leica microwave apparatus (AMW) to dehydrate and infiltrate the Epon resin (The infiltration is done according to the following program, see Table 1).
Table 1. Procedure of resin infiltration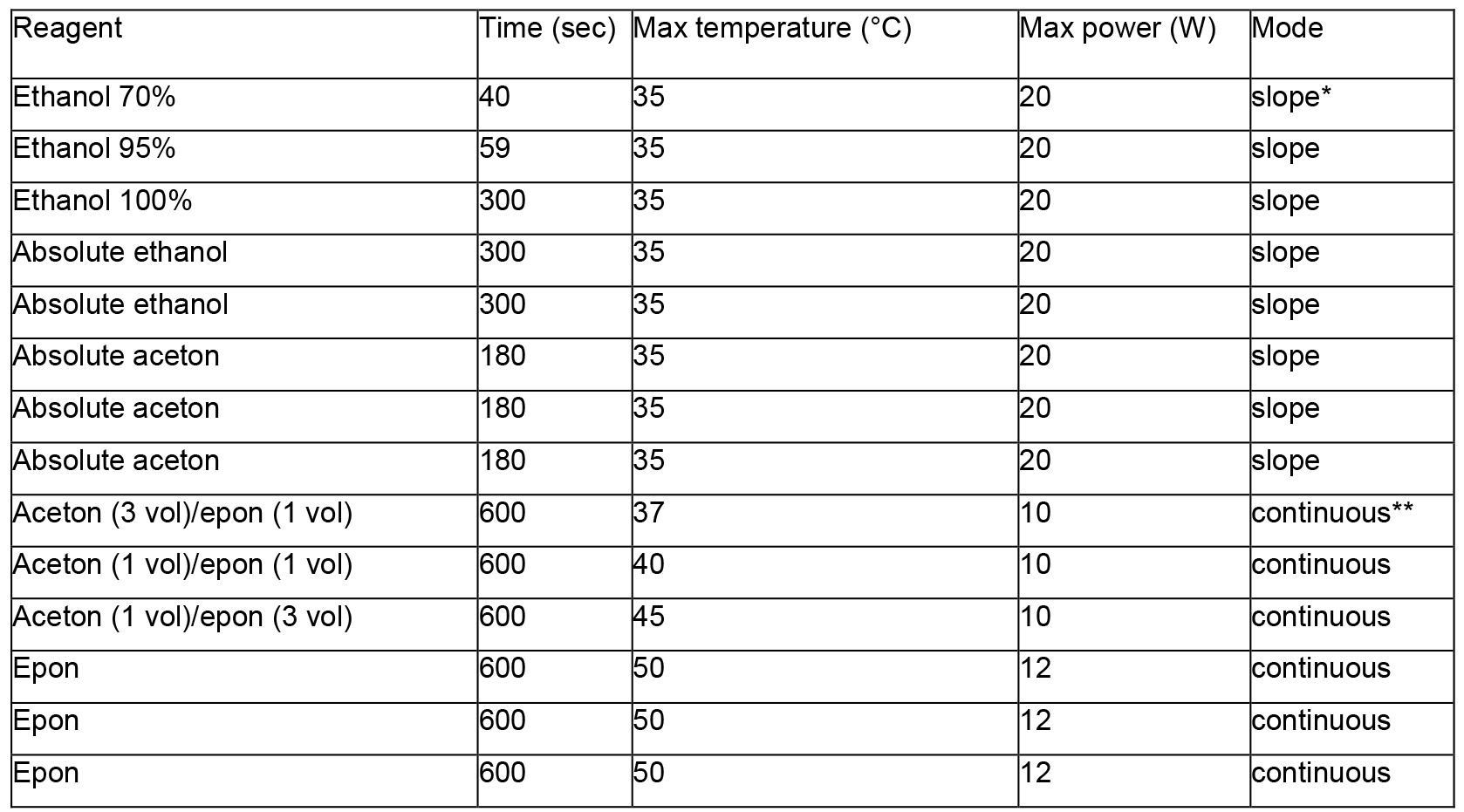
Notes:- *Slope: means that the temperature raised progressively to reach the indicated value at the end of the time.
- **Continuous: means that the temperature reached as soon as possible the indicated value and was then maintained.
- *Slope: means that the temperature raised progressively to reach the indicated value at the end of the time.
- Place the samples in flat bottom embedding capsule filled with pure resin for 48 h of polymerization at 60 °C in an oven.
For optical microscopy- Cut the resin-embedded sample at a thickness of 0.5 µm with a Histo-diamond knife. The sections are placed on a drop of water on a glass slide and warmed to 50 °C on a hot plate to dry and stick on the glass.
- Stain the sections by covering the slide in a 1% borax solution (see Recipe 4) containing 0.1% toluidine blue and 0.2% methylene blue (these two stains were used to contrast thin sections from resin embedded samples). Rinse in water and then cover the slide with an aqueous solution of 0.07% of basic fuchsin (use to stain acidic compounds, see Recipe 5).
- Acquire images (see Figure 3) using an optical microscope (ZEISS, Axioplan 2 imaging) equipped with a CCD color camera (ZEISS, AxioCam MRc)
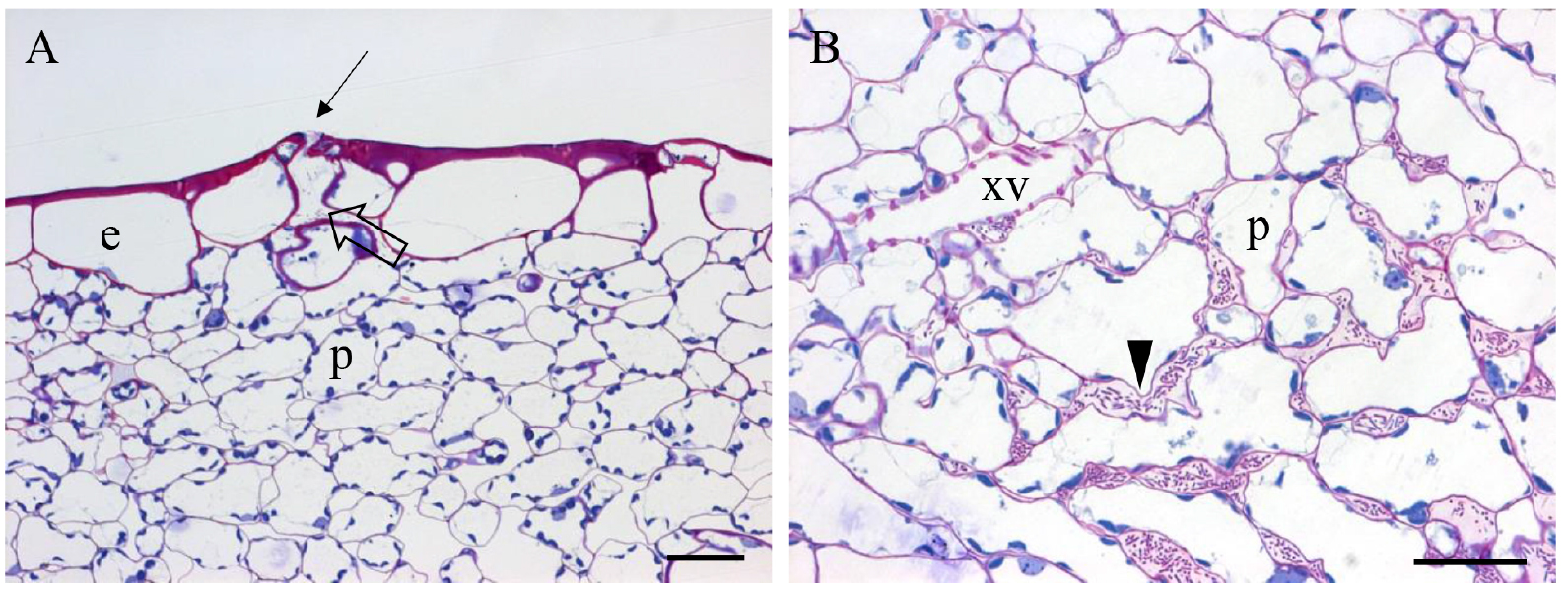
Figure 3. Paradermal sections (0.5 µm in thickness) of resin embedded hydathode (A-B) from cauliflower leaf (3 dpi) infected by Xcc strain 8004::GUS-GFP (Cerutti et al., 2017). e: epidermal cell; p: parenchyma cell of the epithem tissue; xv: xylem vessel. A. Note a pore (arrow), a large chamber below the pore (open arrow); B. The presence of bacteria in the meatuses (arrowhead). Scale bars = 25 µm.
For transmission electron microscopy- Cut the resin-embedded sample at a thickness of 70-80 nm and collect the sections on gold grids and keep to dry overnight before the periodic acid-thiocarbohydrazide-silver (Ag) proteinate (PATAg) treatment.
- Float the sections (on the grids) for 30 min at room temperature on an aqueous solution of 1% (w/v) of periodic acid (see Recipe 6).
- Rinse twice the sections on distilled water for 2 h (1 h for each).
- Treat the sections overnight at 4 °C with a 20% acetic acid solution containing 0.2% thiocarbohydrazide.
- Wash the sections in solutions of decreasing concentrations of acetic acid (20%, 10%, and 5% acetic acid solutions to distilled water) for 30 min each at room temperature.
- Float the sections in a 1% (w/v) silver proteinate (see Recipe 7) for 30 min in the dark at the room temperature.
- Wash the sections in water (2-3 times, 20 min each time) and air-dry.
- Observe the sections under a transmission electron microscope operating at 80 kV and acquire images (see Figure 4).
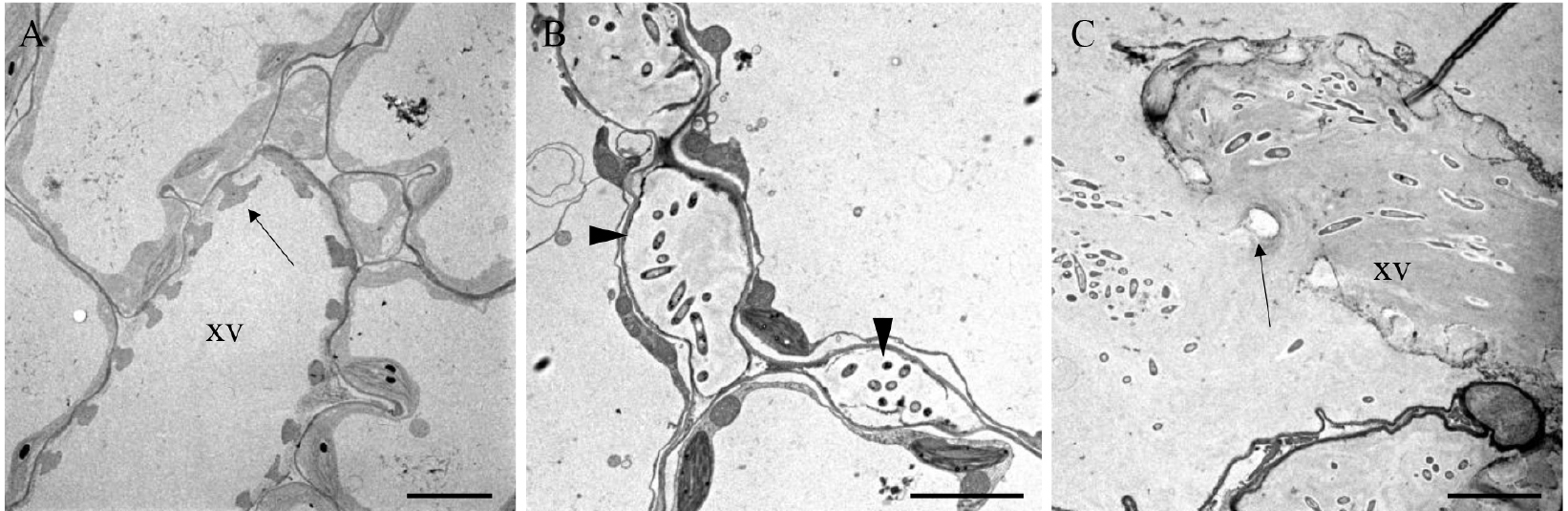
Figure 4. Images of a non-infected hydathode (A) and infected hydathode (B at 3 dpi, and C at 6 dpi) from cauliflower leaf infected by Xcc strain 8004::GUS-GFP (Cerutti et al., 2017). xv: xylem vessel. The arrows indicate the secondary cell wall ornamentation of the xylem vessels. The arrowheads indicate the epithem meatuses colonized by bacteria. Note in (C) the vessel invaded by bacteria. Scale bars = 3 µm.
- With a razor blade, take pieces (2-3 mm2) of leaf margins encompassing hydathodes.
Data analysis
Images of hydathodes from different clarified plant samples can be further analyzed to count the xylem terminations, to determine the size of the different cell types, the number of pores and their size per hydathode. Through analysis of the captured images, we characterized the hydathodes of cauliflower and Arabidopsis (Cerutti et al., 2017). All the measurements were done using Image-Pro Plus 4.0 Imaging software (Media Cybernetics, Silver Spring, MD, USA). It is also possible to use other imaging software like ImageJ:
- To count the xylem terminations (or the pores) of a hydathode, it is most often necessary to acquire images at different focus to visualize all the terminations. Use a measurement tool which saves the positioning of the point feature to avoid overcounting of the xylem terminations within a hydathode. At least 30 to 40 hydathodes were analyzed (Cerutti et al., 2017).
- To determine the size (area) of the cells (epidermal or epithemal cells), it is crucial to obtain well -contrasted images. To solve this problem, stain the cell wall with calcofluor and rinse the samples extensively with water before image acquisition.
Recipes
- Clarification solution
45 g chloral hydrate
7.6 ml distilled water
9.3 ml 60% glycerol - Fixation solution
2.5% glutaraldehyde
0.2 M sodium cacodylate buffer (pH 7.2)
0.1% Triton X-100 - Sodium cacodylate buffer
0.2 M solution of cacodylate (4.28 g in100 ml) in distilled water at pH 7.2
Adjust the pH with several drops of HCl (1 N) - Borax solution with toluidine blue and methylene blue
In an aqueous solution of 1% borax add 0.1% (w/v) of toluidine blue and then 0.2% (w/v) methylene blue - Basic fuchsin solution
0.07% (w/v) of basic fuchsin in distilled water - Periodic acid solution
1% (w/v) of periodic acid in distilled water - Silver proteinate solution
Prepare a fresh solution of 1% (w/v) of silver proteinate in distilled water. In a fume bottle glass, add progressively the silver proteinate under stirring. Filter the solution before use and store it in the dark
Acknowledgments
This work was supported by a PhD grant from the French Ministry of National Education and Research to AC. LIPM is part of the French Laboratory of Excellence project (TULIP ANR-10-LABX-41; ANR-11-IDEX-0002-02). We also thank the Région Occitanie for the financial support of the microscopy platform. Thanks to the CMEAB (Centre de Microscopie Electronique Appliquée à la Biologie) staff for their invaluable technical assistance. The procedures for fixation and embedding were adapted from: a) Glauert, A. M. (1975). Fixation, Dehydration and embedding of biological specimens in Practical Methods in Electron Microscopy. North-Holland Publishing Compagny-Amsterdam. b) Hawes, C. and Satiat-Jeunemaitre, B. (2001). Plant Cell Biology: A Practical Approach. Oxford University Press. The PATAg treatment was adapted from Thiery, J. P. (1967). Mise en évidence des polysaccharides sur coupes fines en microscopie électronique. J; Microsc. 6: 927-1018.
References
- Chen C. C. and Chen Y. R. (2007). Study on laminar hydathodes of Ficus formosana (Moraceae) III. Salt injury of guttation on hydathodes. Bot Stud 48: 215-226.
- Cerutti, A., Jauneau, A., Auriac, M. C., Lauber, E., Martinez, Y., Chiarenza, S., Leonhardt, N., Berthomé, R. and Noël, L. D. (2017). Immunity at cauliflower hydathodes controls systemic infection by Xanthomonas campestris pv campestris. Plant Physiol 174(2): 700-716.
- Glauert, A. M. (1975). Fixation, Dehydration and embedding of biological specimens in Practical Methods in Electron Microscopy. North-Holland Publishing Compagny-Amsterdam.
- Hawes, C. and Satiat-Jeunemaitre, B. (2001). Plant Cell Biology: A Practical Approach. Oxford University Press.
- Thiery, J. P. (1967). Mise en évidence des polysaccharides sur coupes fines en microscopie électronique. J Microsc 6: 927-1018
- Perrin, A. (1972). Contribution à l’étude de l’organisation et du fonctionnement des hydathodes: recherches anatomiques ultrastructurales et physiologiques. Université Claude-Bernard–Lyon.
- Singh, S. (2014). Guttation: quantification, microbiology and implications for phytopathology. In: Lüttg, U., Beyschlag, W., Cushman, J. (Eds). Prog. Bot. Vol. 75. Springer pp 187-214.
- Wang, B., Xu, B., Wang, H., Li, J., Huang, H. and Xu L. (2011). YUCCA genes are expressed in response to leaf adaxial-abaxial juxtaposition and are required for leaf margin development. Plant Physiol 157: 1805-1819.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Cerutti, A., Auriac, M., Noël, L. D. and Jauneau, A. (2017). Histochemical Preparations to Depict the Structure of Cauliflower Leaf Hydathodes. Bio-protocol 7(20): e2452. DOI: 10.21769/BioProtoc.2452.
- Cerutti, A., Jauneau, A., Auriac, M. C., Lauber, E., Martinez, Y., Chiarenza, S., Leonhardt, N., Berthomé, R. and Noël, L. D. (2017). Immunity at cauliflower hydathodes controls systemic infection by Xanthomonas campestris pv campestris. Plant Physiol 174(2): 700-716.
Category
Plant Science > Plant immunity > Host-microbe interactions
Plant Science > Plant cell biology > Cell imaging
Cell Biology > Cell imaging > Fixed-tissue imaging
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link