- Protocols
- Articles and Issues
- About
- Become a Reviewer
Most Read
Most viewed in the last three months
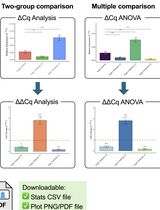
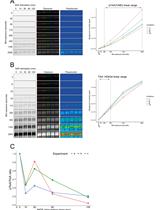
Click-qPCR: A Simple Tool for Interactive qPCR Data Analysis
Real-time quantitative PCR (qPCR) is a pivotal technique for analyzing gene expression and DNA copy number variations. However, the limited availability of user-friendly software tools for qPCR data analysis presents a significant challenge for experimental biologists with limited computational skills. To address this issue, we developed Click-qPCR, a user-friendly and web-based Shiny application for qPCR data analysis. Click-qPCR streamlines ΔCq and ΔΔCq calculations using user-uploaded CSV data files. The interactive interface of the application allows users to select genes and experimental groups and perform Welch’s t tests and one-way analysis of variance with Dunnett’s post-hoc test for pairwise and multi-group comparisons, respectively. Results are visualized via interactive bar plots (mean ± standard deviation with individual data points) and can be downloaded as publication-quality images, along with summary statistics. Click-qPCR empowers researchers to efficiently process, interpret, and visualize qPCR data regardless of their programming experience, thereby facilitating routine analysis tasks. Click-qPCR Shiny application is available at https://kubo-azu.shinyapps.io/Click-qPCR/, while its source code and user guide are available at https://github.com/kubo-azu/Click-qPCR.
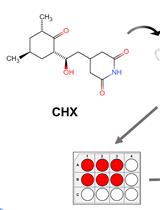
Cycloheximide (CHX) Chase Assay to Examine Protein Half-life
Cycloheximide (CHX) is a small molecule derived from Streptomyces griseus that acts as fungicide. As a ribosome inhibitor, CHX can restrict the translation elongation of eukaryotic protein synthesis. Once protein synthesis is inhibited by CHX, the level of intracellular proteins decreases by degradation through the proteasome or lysosome system. Thus, the CHX chase assay is widely recognized and used to observe intracellular protein degradation and to determine the half-life of a given protein in eukaryotes. Here, we present a complete experimental procedure of the CHX chase assay.Graphical overview
A Guide to Basic RNA Sequencing Data Processing and Transcriptomic Analysis
RNA sequencing (RNA-Seq) has transformed transcriptomic research, enabling researchers to perform large-scale inspection of mRNA levels in living cells. With the growing applicability of this technique to many scientific investigations, the analysis of next-generation sequencing (NGS) data becomes an important yet challenging task, especially for researchers without a bioinformatics background. This protocol offers a beginner-friendly step-by-step guide to analyze NGS data (starting from raw .fastq files), providing the required codes with an explanation of the different steps and software used. We outline a computational workflow that includes quality control, trimming of reads, read alignment to the genome, and gene quantification, ultimately enabling researchers to identify differentially expressed genes and gain insights on mRNA levels. Multiple approaches to visualize this data using statistical and graphical tools in R are also described, allowing the generation of heatmaps and volcano plots to represent genes and gene sets of interest.
Generation of Human Induced Pluripotent Stem Cell (hiPSC)-Derived Astrocytes for Amyotrophic Lateral Sclerosis and Other Neurodegenerative Disease Studies
Astrocytes are increasingly recognized for their important role in neurodegenerative diseases like amyotrophic lateral sclerosis (ALS). In ALS, astrocytes shift from their primary function of providing neuronal homeostatic support towards a reactive and toxic role, which overall contributes to neuronal toxicity and cell death. Currently, our knowledge on these processes is incomplete, and time-efficient and reproducible model systems in a human context are therefore required to understand and therapeutically modulate the toxic astrocytic response for future treatment options. Here, we present an efficient and straightforward protocol to generate human induced pluripotent stem cell (hiPSC)-derived astrocytes implementing a differentiation scheme based on small molecules. Through an initial 25 days, hiPSCs are differentiated into astrocytes, which are matured for 4+ weeks. The hiPSC-derived astrocytes can be cryopreserved at every passage during differentiation and maturation. This provides convenient pauses in the protocol as well as cell banking opportunities, thereby limiting the need to continuously start from hiPSCs. The protocol has already proven valuable in ALS research but can be adapted to any desired research field where astrocytes are of interest.Key features• This protocol requires preexisting experience in hiPSC culturing for a successful outcome.• The protocol relies on a small molecule differentiation scheme and an easy-to-follow methodology, which can be paused at several time points.• The protocol generates >50 × 106 astrocytes per differentiation, which can be cryopreserved at every passage, ensuring a large-scale experimental output.Graphical overview
Generation of 3D Human iPSC-Derived Multi-Cell Type Neurospheres for Studying Neuron, Astrocyte, and Microglia Crosstalk
Three-dimensional (3D) human brain tissue models derived from induced pluripotent stem cells (iPSCs) have transformed the study of neural development and disease in vitro. While cerebral organoids offer high structural complexity, their large size often leads to necrotic core formation, limiting reproducibility and challenging the integration of microglia. Here, we present a detailed, reproducible protocol for generating multi-cell type 3D neurospheres that incorporate neurons, astrocytes, and optionally microglia, all derived from the same iPSCs. While neurons and astrocytes differentiate spontaneously from neural precursor cells, generated by dual SMAD-inhibition (blocking BMP and TGF-b signaling), microglia are generated in parallel and can infiltrate the mature neurosphere tissue after plating neurospheres into 48-well plates. The system supports a range of downstream applications, including functional confocal live imaging of GCaMP6f after adeno-associated virus (AAV) transduction of neurospheres or immunofluorescence staining after fixation. Our approach has been successfully implemented across multiple laboratories, demonstrating its robustness and translational potential for studying neuron–glia interactions and modeling neurodegenerative processes.
A Simple, Reproducible Procedure for Chemiluminescent Western Blot Quantification
Western blotting is a universally used technique to identify specific proteins from a heterogeneous and complex mixture. However, there is no clear and common procedure to quantify the results obtained, resulting in variations due to the different software and protocols used in each laboratory. Here, we have developed a procedure based on the increase in chemiluminescent signal to obtain a representative value for each band to be quantified. Images were processed with ImageJ and subsequently compared using R software. The result is a linear regression model in which we use the slope of the signal increase within the combined linear range of detection to compare between samples. This approach allows to quantify and compare protein levels from different conditions in a simple and reproducible way. Graphical overview
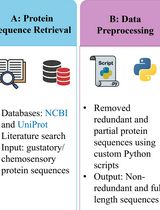
A Step-by-Step Computational Protocol for Functional Annotation and Structural Modelling of Insect Chemosensory Proteins
Insects rely on chemosensory proteins, including gustatory receptors, to detect chemical cues that regulate feeding, mating, and oviposition behaviours. Conventional approaches for studying these proteins are limited by the scarcity of experimentally resolved structures, especially in non-model pest species. Here, we present a reproducible computational protocol for the identification, functional annotation, and structural modelling of insect chemosensory proteins, demonstrated using gustatory receptors from the red palm weevil (Rhynchophorus ferrugineus) as an example. The protocol integrates publicly available sequence data with OmicsBox for functional annotation and ColabFold for high-confidence structure prediction, providing a step-by-step framework that can be applied to genome-derived or transcriptomic datasets. The workflow is designed for broad applicability across insect species and generates structurally reliable protein models suitable for downstream applications such as ligand docking or molecular dynamics simulations. By bridging functional annotation with structural characterisation, this protocol enables reproducible studies of chemosensory proteins in agricultural and ecological contexts and supports the development of novel pest management strategies.
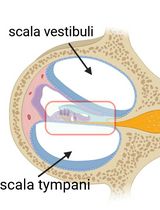
Cochlear Organ Dissection, Immunostaining, and Confocal Imaging in Mice
The organ of Corti, located in the inner ear, is the primary organ responsible for animal hearing. Each hair cell has a V-shaped or U-shaped hair bundle composed of actin-filled stereocilia and a kinocilium supported by true transport microtubules. Damage to these structures due to noise exposure, drug toxicity, aging, or environmental factors can lead to hearing loss and other disorders. The challenge when examining auditory organs is their location within the bony labyrinth and their small and fragile nature. This protocol describes the dissection procedure for the cochlear organ, followed by confocal imaging of immunostained endogenous and fluorescent proteins. This approach can be used to understand hair cell physiology and the molecular mechanisms required for normal hearing.
Room-Temperature Storage of Zebrafish and Medaka Sperm Using Lactic Acid-Stabilized L-15 Medium
Zebrafish offer numerous advantages as a vertebrate model because of their rapid development, high fecundity, transparent embryos, and ease of genetic manipulation. A wide variety of transgenic and mutant fish lines have been generated, and efficiently sharing these resources is crucial for advancing research. Zebrafish lines have typically been exchanged as early embryos, adult fish, or cryopreserved sperm, making transportation costly and logistically challenging. Here, we provide a protocol for preserving functional zebrafish sperm for more than 7 days at room temperature and subsequent in vitro fertilization using the preserved sperm. In this protocol, sperm collected either from the cloaca of an anesthetized male or from dissected testes is stored in L-15-based storage medium. Importantly, the storage medium, originally developed for zebrafish, is also applicable to medaka, another widely used vertebrate model. This sperm storage method allows researchers to ship sperm using low-cost methods and to investigate key factors for motility and fertilizing ability in those sperm.
An Optimized Protocol for Simultaneous Propagation of Patient-derived Organoids and Matching CAFs
Recurrent hormone receptor-positive (HR+) breast cancer is a leading cause of cancer mortality in women. Recurrence and resistance to targeted therapies have been difficult to study due to the long clinical course of the disease, the complex nature of resistance, and the lack of clinically relevant model systems. Existing models are limited to a few HR+ cell lines, organoid models, and patient-derived xenograft models, all lacking components of the human tumor microenvironment. Furthermore, the low take rate and loss of estrogen receptor (ER) expression in patient-derived organoids (PDOs) has been challenging. Our protocol allows simultaneous isolation of PDOs and matching cancer-associated fibroblasts (CAFs) from primary and metastatic HR+ breast cancers. Importantly, our protocol has a higher take rate and enables long-term culturing of PDOs that retain ER expression. Our matching PDOs and CAFs will provide researchers with a new resource to study the influence of the tumor microenvironment on various aspects of cancer biology such as cell growth and drug resistance in HR+ breast cancer.
Rapid and solvent-free, 2-hydroxyethyl methacrylate (HEMA)-acrylamide (AAm) copolymer-based optical clearing of tissue for fluorescent imaging
The study of whole organs or tissues and their cellular components and structures has been historically limited by their natural opacity, which is caused by the optical heterogeneity of the tissue components that scatter light as it traverses through the tissue, making 3D tissue imaging highly challenging. In recent years, tissue clearing techniques have received widespread attention and undergone rapid development. We recently demonstrated the synthesis of a 2-hydroxyethyl methacrylate (HEMA)-acrylamide (AAm) copolymer. This was achieved using antipyrine (ATP) and 2,2′-thiodiethanol (TDE) as solvents. The resulting solution rapidly embedded tissue samples with a high degree of transparency and is compatible with multiple fluorescence labeling techniques. The method exhibits significant transparency effects across a range of organs, comprising the heart, liver, spleen, lung, kidney, brain (whole and sectioned), esophagus, and small intestine. It can enable volumetric imaging of tissue up to the scale of mouse organs, decrease the duration of the clearing, and preserve emission from fluorescent proteins and dyes. To facilitate the use of this powerful tool, we have provided here a detailed step-by-step protocol that should allow any laboratory to use tissue transparency technology to achieve transparency of tissues and organs.
Cloning a Chloroplast Genome in Saccharomyces cerevisiae and Escherichia coli
Chloroplast genomes present an alternative strategy for large-scale engineering of photosynthetic eukaryotes. Prior to our work, the chloroplast genomes of Chlamydomonas reinhardtii (204 kb) and Zea mays (140 kb) had been cloned using bacterial and yeast artificial chromosome (BAC/YAC) libraries, respectively. These methods lack design flexibility as they are reliant upon the random capture of genomic fragments during BAC/YAC library creation; additionally, both demonstrated a low efficiency (≤ 10%) for correct assembly of the genome in yeast. With this in mind, we sought to create a highly flexible and efficient approach for assembling the 117 kb chloroplast genome of Phaeodactylum tricornutum, a photosynthetic marine diatom. Our original article demonstrated a PCR-based approach for cloning the P. tricornutum chloroplast genome that had 90%–100% efficiency when screening as few as 10 yeast colonies following assembly. In this article, we will discuss this approach in greater depth as we believe this technique could be extrapolated to other species, particularly those with a similar chloroplast genome size and architecture.
Characterizing Tissue Oxygen Tension During Neurogenesis in Human Cerebral Organoids
Oxygen tension is a key regulator of early human neurogenesis; however, quantifying intra-tissue O2 in 3D models for an extended period remains difficult. Existing approaches, such as needle-type fiber microsensors and intensity-based oxygen probes or time-domain lifetime imaging, either perturb the organoids or require high excitation doses that limit the measurement period. Here, we present a step-by-step protocol to measure intra-organoid oxygen in human cerebral organoids (hCOs) using embedded ruthenium-based CPOx microbeads and widefield frequency-domain fluorescence lifetime imaging microscopy (FD-FLIM). The workflow covers dorsal/ventral cerebral organoid patterning, organoid fusion at day 12 with co-embedded CPOx beads, standardized FD-FLIM acquisition (470-nm external modulation, 16 phases at 50 kHz, dual-tap camera), automated bead detection and lifetime extraction in MATLAB, and session-matched Stern–Volmer calibration with Ru(dpp)3(ClO4)2 to convert lifetimes to oxygen concentration. The protocol outputs per-bead oxygen maps and longitudinal patterns stratified by bead location (intra-organoid vs. gel) and sample state (healthy vs. abnormal), enabling direct linkage between developmental growth and oxygen dynamics.
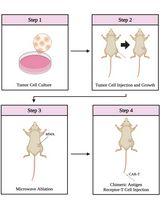
Combining Microwave Ablation With CAR-T-Cell Therapy in Tumor-Bearing Mouse Models
Microwave ablation (MWA) is a thermal ablation technique widely used for local tumor control that has the added potential to stimulate systemic anti-tumor immunity. Although MWA alone rarely eliminates recurrent or metastatic disease, its ability to remodel the tumor microenvironment makes it a promising partner for adoptive cell therapies such as chimeric antigen receptor (CAR)-T cells. However, reproducible protocols for combining these approaches remain limited. This protocol describes the integration of MWA with CAR-T therapy in tumor-bearing mouse models. Human hepatocellular carcinoma cell lines (Hep3B and SK-HEP-1) are inoculated subcutaneously into NOG mice to establish tumors. Localized MWA is performed at adjustable power and duration to induce partial or complete ablation. At defined intervals following MWA, CAR-T cells derived from healthy donor T cells and transduced with a lentiviral vector are injected intravenously. This experimental design uniquely separates MWA and CAR-T delivery, enabling precise evaluation of thermal preconditioning effects on the tumor microenvironment and subsequent CAR-T activity. By combining localized ablation with adoptive immunotherapy, the protocol provides a translationally relevant platform to optimize treatment timing, enhance CAR-T efficacy in solid tumors, and address key barriers in tumor immunology and cancer therapy.
Live-Cell Monitoring of Piecemeal Chloroplast Autophagy
When plants undergo senescence or experience carbon starvation, leaf cells degrade proteins in the chloroplasts on a massive scale via autophagy, an evolutionarily conserved process in which intracellular components are transported to the vacuole for degradation to facilitate nutrient recycling. Nonetheless, how portions of chloroplasts are released from the main chloroplast body and mobilized to the vacuole remains unclear. Here, we developed a method to observe the autophagic transport of chloroplast proteins in real time using confocal laser-scanning microscopy on transgenic plants expressing fluorescently labeled chloroplast components and autophagy-associated membranes. This protocol enabled us to track changes in chloroplast morphology during chloroplast-targeted autophagy on a timescale of seconds, and it could be adapted to monitor the dynamics of other intracellular processes in plant leaves.
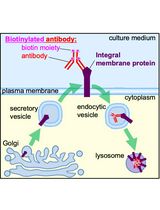
Monitoring Endocytosis of Integral Membrane Proteins Using Western Blot-Based Detection of Biotinylated Antibody Uptake
The antibody-uptake assay is a commonly used technique to monitor endocytosis of integral membrane proteins including transmembrane and glycosylphosphatidylinositol-anchored proteins (GPI-APs). The antibody-uptake assay typically involves incubating live cells with fluorophore-conjugated antibodies directed against the extracellular domain of the integral membrane protein of interest. Antibody uptake is then detected by flow cytometry or confocal microscopy. However, these detection modalities may be inaccessible to some labs or require extensive training to operate. Thus, we developed an easy and novel sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and western blot-based approach to the antibody-uptake assay that exploits the strong affinity between biotin and streptavidin. Instead of incubating cells with fluorophore-conjugated antibodies to monitor antibody uptake, our assay involves incubating cells with biotinylated antibodies, processing the cell lysates for western blot, and probing the membrane with detectably conjugated streptavidin. From preparation to quantification, this protocol requires less hands-on time than other approaches and is amenable to small-scale drug or siRNA screens. Here, we demonstrate the utility of our approach using the well-characterized misfolded GPI-AP, YFP-tagged C179A mutant of prion protein (YFP-PrP*), as our model substrate. YFP-PrP* constitutively traffics to the plasma membrane (PM), where it binds to anti-GFP antibody, and immediately undergoes endocytosis to lysosomes. To validate our protocol, we present measurements of antibody uptake under conditions known to enhance or inhibit YFP-PrP*’s traffic to the PM. Using this assay, we present new evidence that, under certain conditions, YFP-PrP* is able to undergo degradation via a pathway that does not involve exposure on the cell surface.
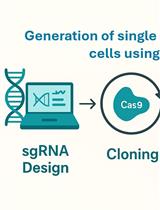
Protocol for Generation of Single-Gene Knockout in Hard-to-Transfect THP1 Cell Lines Using CRISPR/Cas9
This protocol provides a step-by-step approach for generating single-gene knockout in hard-to-transfect suspension immune cell lines like THP1, specifically demonstrated by knocking out the GSDMD gene. By employing CRISPR-Cas9 system delivered via lentivirus, this protocol enables precise gene disruption through targeted single-guide RNAs (sgRNAs). Key steps include designing specific sgRNAs, cloning them into a CRISPR vector, viral packaging, and transducing the target cells, followed by selection and validation. This optimized protocol is particularly useful for functional studies in immune cells, allowing researchers to reliably explore gene function in complex cellular pathways.
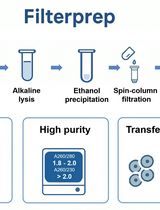
Plasmid DNA Purification Using Filterprep With an Optional Endotoxin Removal Step
This protocol presents a modified version of the Filterprep method originally reported in New Biotechnology, adding an optional step to reduce endotoxin levels. Filterprep is a simple, rapid, and cost-effective approach to plasmid DNA purification that couples ethanol precipitation with a single spin-column filtration step, eliminating chaotropic salts and silica binding. The formulations and parameters are fully transparent and do not rely on proprietary buffers, using only standard laboratory reagents and widely available miniprep columns. Under matched conditions, the method recovers high-purity plasmid DNA with yields up to fivefold higher than those obtained with representative commercial midiprep kits. The workflow is readily adoptable in most molecular biology laboratories and, under routine conditions, can be completed in approximately 40 min. The resulting DNA is suitable for molecular cloning, PCR, sequencing, and other downstream biochemical applications. Endotoxin is a lipopolysaccharide (LPS) found in the outer membrane of Gram-negative bacteria and may carry over during plasmid preparation. For experiments requiring lower endotoxin input, an optional modification resuspends the DNA pellet in a Triton X-114 wash buffer before column loading to decrease lipopolysaccharide carryover. The method is modular and extensible, allowing adjustment of precipitation and wash conditions, variation in the number of washes, selection of alternative column formats, and integration of endotoxin-reduction modules without altering the core principle. These features facilitate troubleshooting and quality control, enable scaling from routine batches to larger culture volumes and higher throughput, and allow seamless integration with existing workflows.
In Vitro Bone Marrow–Derived Dendritic Cells (BMDC) Generation for Antigen Presentation Assay
Dendritic cells (DC) are sentinel cells of the immune system that process and present antigens to activate T cells, thus serving to bridge the innate and adaptive immune systems. DCs are particularly efficient at cross-presentation whereby exogenously acquired antigens are processed and presented in context with MHCI molecules to activate CD8+ T cells. Assaying antigen presentation by DCs is a critical parameter in assessing immune functionality. However, the low abundance of bona fide DCs within the lymphoid compartments limits the utility of such assays. An alternative approach employing the culturing of bone marrow cells in the presence of factors needed for DC lineage commitment can result in the differentiation of bone marrow dendritic cells (BMDCs). This protocol details the process of in vitro generation of BMDCs and demonstrates their subsequent utility in antigen presentation assays. The protocol described can be adapted to various conditions and antigens.
Quantitative Analysis of the Arabidopsis Leaf Secretory Proteome via TMT-Based Mass Spectrometry
In plants, the apoplast contains a diverse set of proteins that underpin mechanisms for maintaining cell homeostasis, cell wall remodeling, cell signaling, and pathogen defense. Apoplast protein composition is highly regulated, primarily through the control of secretory traffic in response to endogenous and environmental factors. Dynamic changes in apoplast proteome facilitate plant survival in a changing climate. Even so, the apoplast proteome profiles in plants remain poorly characterized due to technological limitations. Recent progress in quantitative proteomics has significantly advanced the resolution of proteomic profiling in mammalian systems and has the potential for application in plant systems. In this protocol, we provide a detailed and efficient protocol for tandem mass tag (TMT)-based quantitative analysis of Arabidopsis thaliana secretory proteome to resolve dynamic changes in leaf apoplast proteome profiles. The protocol employs apoplast flush collection followed by protein cleaning using filter-aided sample preparation (FASP), protein digestion, TMT-labeling of peptides, and mass spectrometry (MS) analysis. Subsequent data analysis for peptide detection and quantification uses Proteome Discoverer software (PD) 3.0. Additionally, we have incorporated in silico–generated spectral libraries using PD 3.0, which enables rapid and efficient analysis of proteomic data. Our optimized protocol offers a robust framework for quantitative secretory proteomic analysis in plants, with potential applications in functional proteomics and the study of trafficking systems that impact plant growth, survival, and health.
Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics