Improve Research Reproducibility A Bio-protocol resource
Improve Research Reproducibility A Bio-protocol resource
Advanced Search
Preprint
Spinning-disc confocal and 2P3A-DSLM imaging
Last updated date: Mar 27, 2021 Views: 1173 Forks: 0
Detailed protocol for spinning-disc confocal imaging
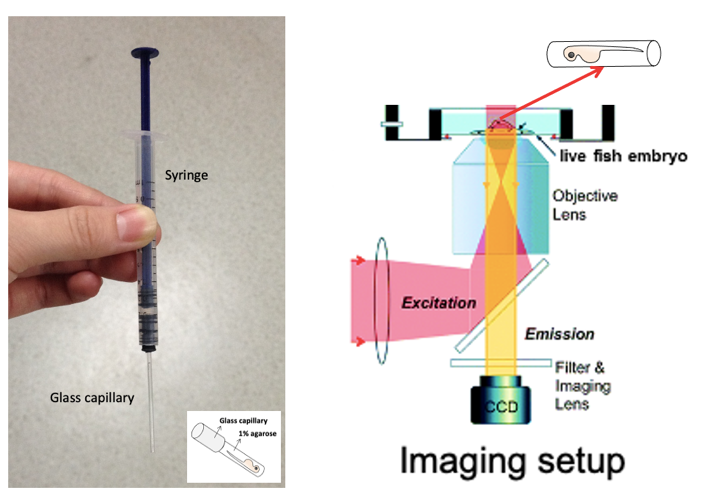
- For time-lapse imaging, images were captured using a commercial Olympus spinning-disc confocal microscope. Due to the limitation of the working distance of our imaging objectives, in this experiment a 10×/0.4 objective was used to ensure that samples (ins: Rcamp signals in living zebrafish embryos) will be illuminated on proper focal plane. During imaging, 1.6 × preamplifier was used for image magnification. Alternatively, “zoom in” of images is also recommended.
- Transferring zebrafish embryo to the agarose: Heat up one aliquot of 1% ultra-pure agarose (Invitrogen) in an EP tube and keep it warm in a heat block set to 38-40℃. Supplement tricaine to a final concentration of 0.01% into the agarose. Select one dechorionated zebrafish embryo and transfer it into the preheated 1% ultra-pure agarose. Try to transfer as little E3 medium as possible.
- Mounting zebrafish embryo: Take up the embryo with a self-designed mounting device (Fig.1) with syringe-attached glass capillary (inner diameter: 0.8-1mm). Make sure to fill with some agarose before taking up the fish embryo into the capillary (avoid bubbles inside) and try to place the embryo positioned close to the end of the capillary and with the embryo head pointing towards the opening of the capillary. Keep the capillary horizontal to avoid leakage of its content until the agarose becomes fully solid. Plug out an agarose cylinder containing the zebrafish embryo and place it in an imaging chamber. Orient the position of the embryo with right side of fish body facing to the bottom of the chamber and the objective (this allows light penetration through the fish embryo and effective illumination of the ins: Rcamp signals).
- Fill the imaging chamber with 200ul basal E3 medium containing 0.01% tricaine to ensure fully cover the cylinder containing the zebrafish embryo.
- Find fish embryo under microscope and image ins: Rcamp signals on the best focal plane. Capture images under basal E3 medium (containing 0.01% tricaine) for 10-15min as the basal calcium signals. Add equal volume (200ul) of 40mM glucose stock solution (in E3 medium with 0.01% tricaine) to the imaging chamber to reach a final concentration of 20mM during stimulation. Capture images under the high glucose E3 medium for 15-30min as the stimulated calcium signals. Alternatively, a perifusion chamber is more preferable for applying glucose or other stimuli stimulation if available.
- Time-lapse images are captured once per second with a 100ms exposure time. Images were collected using MetaMorph software and analyzed with Fiji software.
Detailed protocol for 2P3A-DSLM imaging procedure
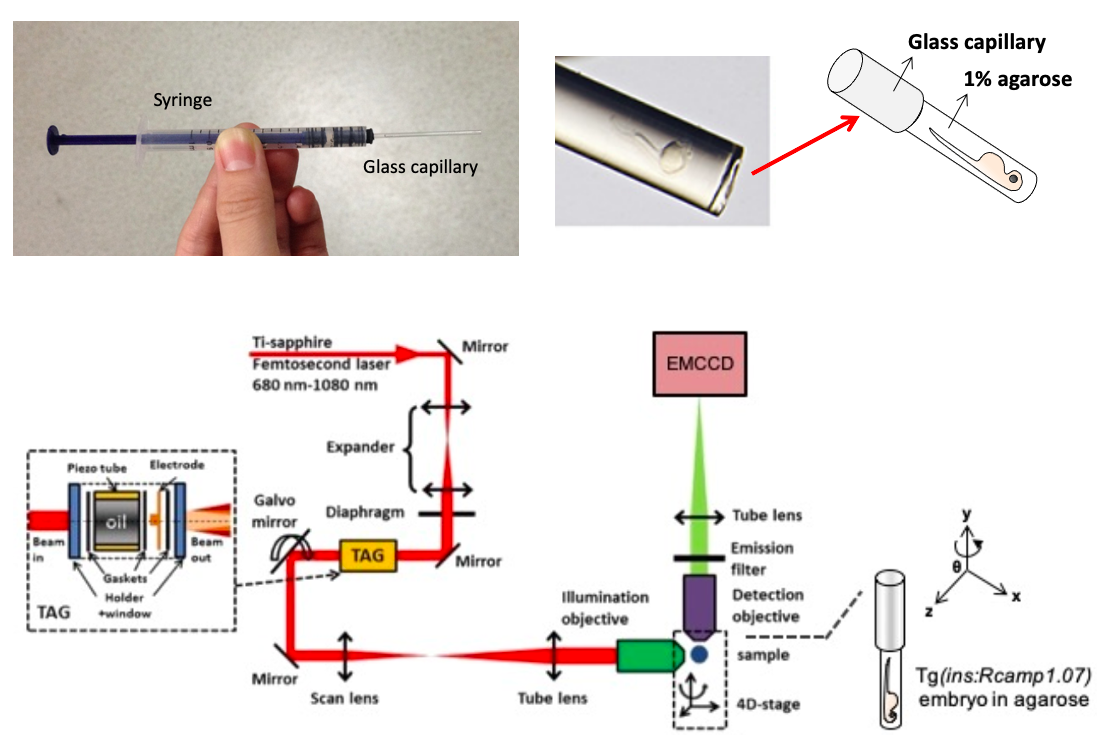
- Images were captured with a home-made 2P3A-DSLM equipped with two 40×/0.8 water lenses, as previously described (Zong et al., 2014, Cell Research).
- Transferring zebrafish embryo to the agarose: Heat up one aliquot of 1% ultra-pure agarose (Invitrogen) in an EP tube and keep it warm in a heat block set to 38-40℃. Supplement tricaine to a final concentration of 0.01% into the agarose. Select one dechorionated zebrafish embryo and transfer it into the preheated 1% ultra-pure agarose. Try to transfer as little additional E3 medium as possible.
- Mounting zebrafish embryo: Take up the embryo with a self-designed mounting device (Fig.2) with syringe-attached glass capillary (inner diameter: 0.8-1mm). Make sure to fill with some agarose before taking up the fish embryo into the capillary (avoid bubbles inside) and try to place the embryo positioned close to the end of the capillary and with the embryo head pointing towards the opening of the capillary. Keep the capillary horizontal to avoid leakage of its content until the agarose becomes fully solid.
- Partially plug out an agarose cylinder containing the zebrafish embryo, locate it vertically using a device holder and immerse in the light-sheet cubic imaging chamber pre-filled with 49ml basal E3 medium containing 0.01% tricaine (Fig.2).
- Rotate the syringe device to orient the embedded embryo with right side of fish body facing to the illumination objective of light sheet microscope (this allows light penetration through the fish embryo and effective illumination of the ins: Rcamp signals).
- Find fish embryo under microscope and image ins: Rcamp signals on the best focal plane. Capture images under basal E3 medium (containing 0.01% tricaine) as the basal calcium signals. Add 1ml of 1M glucose stock solution (in E3 medium with 0.01% tricaine) to the cubic imaging chamber to reach a final concentration of 20mM during stimulation. Capture images under the high glucose E3 medium as the stimulated calcium signals.
- For the 2D time-lapse imaging experiments used in the statistical analysis, the islet was optically sectioned into 5-6 layers (z-step = 10um) to ensure that the calcium transients were recorded in all β-cells within the islet. Each layer was captured with 150ms exposure time. With this strategy, it means 150ms time for imaging each layer and a total time of approx. 1.5s per imaging cycle for imaging the entire islet.
- For the fast volumetric imaging and reconstruction of calcium transients within the whole islet, the islet was optically sectioned into 25 layers (z-step = 2um). Each layer was captured five times with an 8ms exposure time and was averaged as one single image (the averaging process will significantly increase image contrast by reducing noise). With this strategy, it means 40ms time for imaging each layer and a total time of 1s per imaging cycle for imaging the entire islet.
- Images were collected using the HCImage software (Hamamatsu) and processed with R-L deconvolution by the Fiji software. The volumetric calcium transients were reconstructed using Amira software (FEI).
Copyright: Content may be subjected to copyright.
How to cite:
Readers should cite both the Bio-protocol preprint and the original research article where this protocol was used:
- Zhao, J, Liu, Y and Chen, L(2021). Spinning-disc confocal and 2P3A-DSLM imaging. Bio-protocol Preprint. bio-protocol.org/prep978.
- Zhao, J., Zong, W., Zhao, Y., Gou, D., Liang, S., Shen, J., Wu, Y., Zheng, X., Wu, R., Wang, X., Niu, F., Wang, A., Zhang, Y., Xiong, J., Chen, L. and Liu, Y.(2019). In vivo imaging of β-cell function reveals glucose-mediated heterogeneity of β-cell functional development. eLife. DOI: 10.7554/eLife.41540
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
0 Q&A
Spinning
This protocol preprint was submitted via the "Request
a Protocol" track.
Share
Bluesky
X
Copy link
Cancel

