Clone our analysis repository:
git clone https://github.com/sirusb/2CLike_analysis.git
cd 2CLike_analysis/Mapping/BulkRNASeq/
Prepare the genomic annotation following the protocol described here.
Prepare the meta-genome annotation following the protocol described here.
Dowload the Bulk RNA-seq fastq files from GSE133232.
Make sure that the pair-end fastq files of each sample are in a separate directory. In order for the script to work smoothly the directories should be named as shown bellow. Edit the variable "RootDir" to point to the path of the main folder that contains the fastq samples.

If you made any changes, make sure to edit the "branches" variable in our script: Mapping/BulkRNASeq/RNASeq_workflow.groovy
Check that the sample paths are correct by running:
bpipe test RNASeq_workflow.groovy
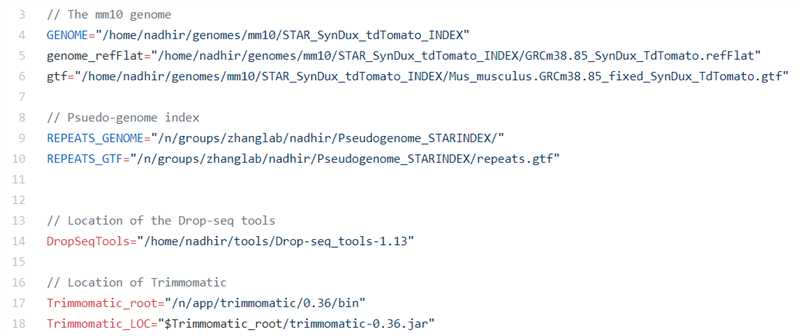
Edit the different software paths and the location to the previously generated .gtf and STAR genomic indexes in the file Mapping/BulkRNASeq/RNASeq_workflow.groovy
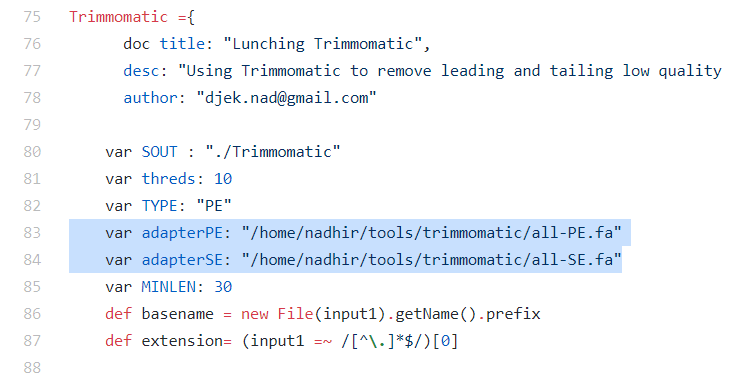
In the Trimmomatic analysis block, make sure to put the correct path the Trimommatics .fa files that contain the adapter sequences.
You can then run the pipeline using bpipe as follows:
bpipe run -r RNASeq_workflow.groovy
If you had any error during any intermediate stage (eg: software path not correct), you can fix it as run "bpipe retry" so the pipeline will continue for the most recent successful stage.
The analysis script used for bulk RNA-seq differential gene expression analysis can be found in our gihub custom script at: 2CLike_analysis/Analysis/bulkRNA_analysis.Rmd
It is advised to open Rstudio and run the script step-by-step and edit the different paths.
Before running the script, make sure that you have the following packages installed:
BiocManager::install(c("edgeR","ggplot2","gridExtra","eulerr","VennDiagram","rtracklayer","RColorBrewer","ggsci","ggrepel")
Make sure to put the correct path to the folder containing the gene expression results generated by the previous script in the variable rootDir.
rootDir <- "D:/Projects/Dux_project/Data/BulkRNASeq"
You can then run the script step-by-step in Rstudio.



