Advanced Search
ATAC sequencing protocol starting from frozen cells
Last updated date: Dec 22, 2020 Views: 975 Forks: 0
ATAC-seq protocol starting from frozen cells
Adapted from Corces, M. et al. Nat Methods 14, 959–962 (2017). https://doi.org/10.1038/nmeth.4396
ATAC-seq from frozen cells – sample requirements
- Bulk ATAC-seq requires each sample to be present as a replicate. Triplicates are better.
- Cells for ATAC-seq should be cryopreserved at high viability. Please ensure that the cells are cryopreserved properly in freezing medium.
- Please send a minimum of 1 million cells per sample.
- Please ship the cells in dry ice.
- Our address is:
Advanced Technology Research Facility
8560 Progress Drive, Room D3043
Frederick, MD 21701
- If you have not already done that, please put in a NAS request for your project. Please select Sequencing Facility – Illumina (CCR) as the service area.
(https://ncifrederick.cancer.gov/Services/Accessioning/Requests/LabServices/New?key=b715aa94-2d8a-474b-b99f-833d894caa2b&stepIndex=0)
List of reagents
- DNase (Worthington cat# LS002007).
- Hanks Balanced Salt Solution.
- PBS.
- 1M Tris-HCl pH 7.4
- 5M NaCl.
- 1M MgCl2.
- Digitonin (Promega cat# G9441) Digitonin is supplied at 2% in DMSO. Dilute 1:1 with water to make a 1% (100x) stock solution. Avoid more than 5 freeze thaw cycles. Can be kept at -20°C for up to 6 months.
- Tween-20 (Sigma/Roche cat# 11332465001) Tween-20 is supplied at 10%. Use at this concentration (100x stock). Store at 4°C.
- NP40 (Sigma/Roche cat# 11332473001) NP40 is supplied at 10%. Use at this concentration (100x stock). Store at 4°C.
- 2x TD buffer (Illumina cat# 15027866).
- Transposase (Illumina cat# 15027865).
- Zymo DNA Clean and Concentrator-5 Kit (cat# D4014).
- NEBNext 2x MasterMix.
Buffer preparation for ATAC-seq protocol (day 1)
1. Prepare base buffer ATAC-RSB (ATAC-Resuspension Buffer, can be stored at room temperature for months).
| Reagent | Final Concentration | Volume for 50 ml |
| 1M Tris-HCl pH 7.4 | 10 mM | 500 ul |
| 5M NaCl | 10 mM | 100 ul |
| 1M MgCl2 | 3 mM | 150 ul |
| Sterile water | NA | 49.25 ml |
2. Prepare DNase stock solution as follows:
2a. Resuspend the lyophilized DNase powder (100 mg) in 15 ml of Hanks Balanced Salt Solution (HBSS). This corresponds to 6.66 mg/ml. 1 mg of DNase corresponds to 2000 Units so 6.66 mg/ml corresponds to 13333 Units/ml.
2b. Aliquot the stock solution and keep them at 4°C up to 3 weeks. Otherwise keep them at -20°C.
2c. Final concentration in the ATAC-Seq sample is 200 Units/ml.
3. Prepare cold ATAC-RSB 1 (70-100ul per sample, prepare on the day of the experiment): Add the three detergents (from the 100X stocks, as described above) to the base ATAC-RSB to final 1X concentration of each. Thus, final concentrations for each: 0.1% NP40, 0.1% Tween-20, and 0.01% Digitonin.
4. Prepare cold wash-out buffer / ATAC-RSB 2 (1 ml per sample, prepare on the day of the experiment): Add Tween-20 to the base ATAC-RSB to 1X final concentration (= 0.1% Tween-20 final).
5. Prepare Transposition mix, during step 30 in the ATAC-seq protocol section, as follows (for 1 reaction):
25 ul 2x TD buffer,
2.5 ul transposase (100nM final),
16.5 ul PBS,
0.5 ul 1% digitonin,
0.5 ul 10% Tween-20,
5 ul H2O.
Cell Preparation for ATAC-seq (day 1)
6. Warm up normal media (IMDM + 10% FBS) at 37°C.
7. Add 4 ml of warm media into a 15 ml tube (1 tube per sample).
8. Remove cryopreserved cell vials from -80°C and place them into a bucket with ice.
9. Place the cryopreserved cells vials at 37°C (no more than 3 vials at a time).
10. When cells are half thawed (typically 1-2 min), add half volume of warm media inside the vial.
11. Pipette up and down several times until cells are completely thawed.
12. Immediately transfer the cells into the 15 ml tube with warm media.
13. Spin down cells at 500g, room temperature, 5 min.
14. Discard supernatant and resuspend the cell pellet with 1 ml of 37°C media.
15. Transfer the resuspended cells to a fresh eppendorf.
16. Add 15 ul DNase from the aliquoted stock solution and incubate for 30 min at 37°C.
17. Spin down cells at 500g, 4°C, 5 min.
18. Discard supernatant and resuspend the cell pellet with 1 ml of ice cooled PBS (first PBS wash).
19. Spin down cells at 500g, 4°C, 5 min.
20. Repeat last two steps again (second PBS wash).
21. Using the cell number from the customer, resuspend the cell pellet with a PBS volume to have an approximate concentration of 1-5 million/ml cells.
22. Count cells and viability with the cell counter.
ATAC-seq protocol (day 1)
23. Pellet 50,000 viable cells at 500 RCF at 4°C for 5 min in a fixed angle centrifuge.
24. Aspirate all supernatant, carefully avoiding visible (sometimes invisible) cell pellet, using two pipetting steps (aspirate down to 100 ul with a p1000 pipette and remove final 100 ul with a p200 pipette).
25. Add 50 ul cold ATAC-Resuspension Buffer (RSB) 1 containing 0.1% NP40, 0.1% Tween-20, and 0.01% Digitonin and pipette up and down 3 times.
26. Incubate on ice for 3 minutes.
27. Wash out lysis with 1 ml of cold ATAC-RSB 2 containing 0.1% Tween-20 but NO NP40 or digitonin and invert tube 3 times to mix.
28. Pellet nuclei at 500 RCF for 10 min at 4°C in a fixed angle centrifuge.
29. Aspirate all supernatant, carefully avoiding visible cell pellet, using two pipetting steps (aspirate down to 100 ul with a p1000 pipette and remove final 100 ul with a p200 pipette).
30. Resuspend cell pellet in 50 ul of transposition mix by pipetting up and down 6 times. Transposition mix = (25 ul 2x TD buffer, 2.5 ul transposase (100nM final), 16.5 ul PBS, 0.5 ul 1% digitonin, 0.5 ul 10% Tween-20, 5 ul H2O).
31. Incubate reaction at 37°C for 30 minutes in a thermomixer with 1000 RPM mixing.
32. Clean up the reaction with a Zymo DNA Clean and Concentrator-5 Kit according with the manufacturer instructions. Elute DNA in 21 ul Elution Buffer and save at -20°C.
Library preparation (day 2)
33. Amplify transposed DNA for 9 cycles using NEBNext 2x MasterMix.
| 25 uM Primer Ad1 | 2.5 ul |
| 25 uM Primer Ad2 | 2.5 ul |
| 2x NEBNext Master Mix | 25 ul |
| Transposed DNA sample | 20 ul |
| Total volume | 50 ul |
| Cycling conditions | |
| 72°C | 5 min |
| 98°C | 30 sec |
| Then 9 cycles of | |
| 98°C | 10 sec |
| 63°C | 30 sec |
| 72°C | 1 min |
| Hold at 4°C | |
Double-sided beads purification (to remove primer dimers and fragments over 1000bp) (day 2)
34. Add 0.5X volume (25 ul) SPRI beads to the PCR samples and pipet up and down 10 times to mix thoroughly.
35. Incubate 10 min at room temperature.
36. Place PCR tubes in a magnetic rack for 5 min or till supernatant is clear.
37. Transfer supernatant to a new PCR tube.
38. Add 1.3X original volume (65 ul) SPRI beads and pipette up and down 10 times to mix thoroughly.
39. Incubate 10 min at room temperature.
40. Place PCR tubes in a magnetic rack for 5 min or till supernatant is clear.
41. Discard supernatant.
42. Wash beads twice with 200 ul of 80% Ethanol (freshly made).
43. Eliminate ethanol traces by spinning down the tubes for 5 sec, placing them back in a magnetic rack and aspirating the ethanol with a p10/p20 tip.
44. Resuspend beads in 20 ul nuclease-free H2O.
45. Incubate 1 min at room temperature.
46. Place PCR tubes in a magnetic rack for 5 min.
47. Transfer supernatant to a new tube.
48. Run 0.5 ul with High Sensitivity Chip Bioanalyzer.
49. Save sample at -20°C.
Depending on the Bioanalyzer profile more PCR cycles may be required (non or very low amplification). Also, in many experiments a second beads purification may be necessary to further remove primer dimers (1X) and/or fragments over 1000 bp (repeat double size selection).
ATAC-Seq oligos for PCR (indexes)
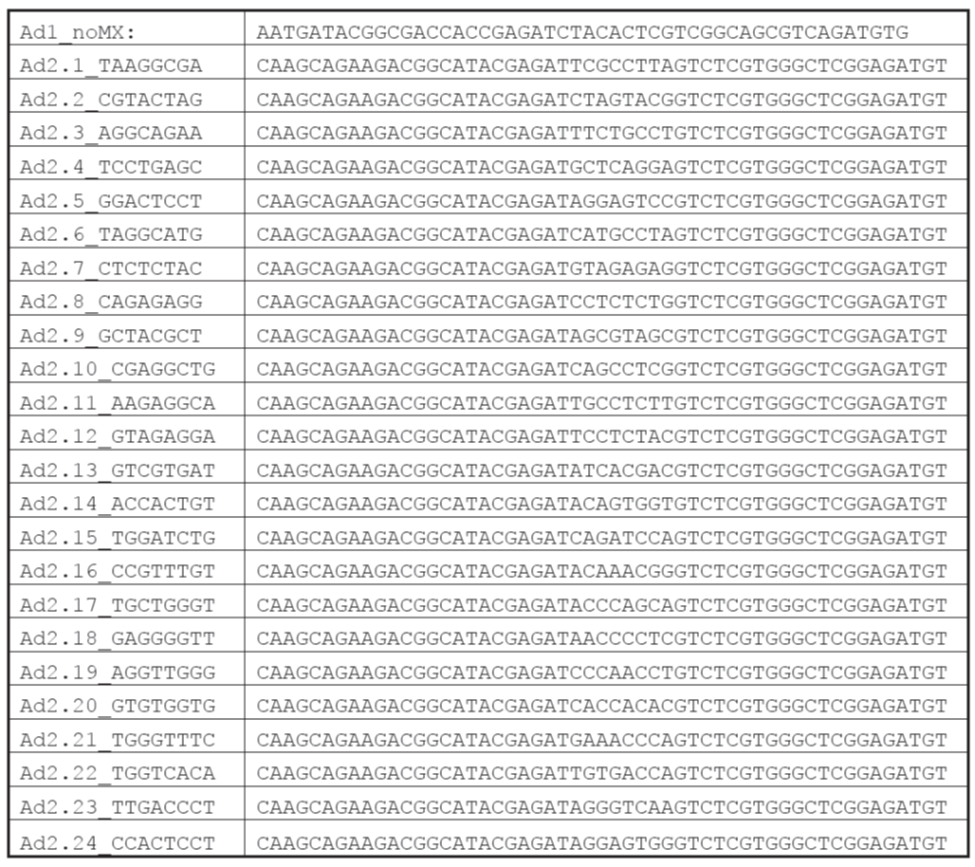
Order the oligos from IDT. These oligos will come lyophilized. Resuspend them in nuclease-free water to make a stock of 100 uM. Finally, from the 100 uM stock make a second stock with nuclease-free water at 25 uM. Save both stocks at -20°C.
- Mehta, M and Caravaca-Guasch, J M(2020). ATAC sequencing protocol starting from frozen cells. Bio-protocol Preprint. bio-protocol.org/prep717.
- Krishna, S., Lowery, F. J., Copeland, A. R., Bahadiroglu, E., Mukherjee, R., Jia, L., Anibal, J. T., Sachs, A., Adebola, S. O., Gurusamy, D., Yu, Z., Hill, V., Gartner, J. J., Li, Y. F., Parkhurst, M., Paria, B., Kvistborg, P., Kelly, M. C., Goff, S. L., Altan-Bonnet, G., Robbins, P. F. and Rosenberg, S. A.(2020). Stem-like CD8 T cells mediate response of adoptive cell immunotherapy against human cancer. Science 370(6522). DOI: 10.1126/science.abb9847
Category
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link

