Advanced Search
Protocols for FACS purification of cardiac total interstitial single cells from adult mouse cardiac ventricles
Last updated date: Nov 9, 2020 Views: 1251 Forks: 0
Abstract:
Cardiomyocytes are the main functional cell types of the heart. However beside the cardiomyocytes, the heart also contains large population of diverse cell types, including fibroblasts, vascular, immune and nerve cells that play crucial roles in cardiac hemostasis and repair. Those non-myocytes cells we collectively refer to as the total interstitial cell population (TIP). Here, we describe a protocol for isolation and purification of high-quality, live, single TIP cells from whole mouse hearts or dissected ventricles using Fluorescent Activated Cell Sorting (FACS). Please refer to Farbehi et al 2019 on the execution of this protocol on dissected ventricles from healthy and infarcted hearts of C57BL/6J and PdgfraGFP/+ mice (1). This article describes a protocol for isolation and FACS sorting of live TIP cells from C57BL/6J mice. To isolate specific cell types within TIP, FACS gating for genetic lineage tags such as GFP, or staining with cell-type specific, fluorophore-conjugated antibodies needs to be performed.
Note: This protocol is adapted from a previous protocol (2).
Keyword: Total interstitial population, Adult heart
Resource table(materials, reagents, and equipment):
| Reagents or instruments | company | Catalogue number |
| Bovine Serum Albumin (BSA)1X pH 7.4 | Sigma-Aldrich | A2153 |
| Fetal Bovine Serum (FBS) | Thermo Fisher Scientific | 10100 |
| Red Blood Cell (RBC) Lysis Buffer | Sigma-Aldrich | R7757 |
| Dead Cell Removal MicroBeads | Miltenyi Biotec | 130-090-101 |
| Collagenase type II | Worthington Biochemical Corporation | LS004177 |
| DAPI | Thermo Fisher Scientific | D1306 |
| Refrigerated benchtop centrifuge for 5, 15, 50 mL tubes | - | - |
| Refrigerated benchtop microcentrifuge for 1.5-2 mL Eppendorf | - | - |
| DNA LoBind 1.5 mL tubes | Eppendorf | 022431005 |
| 6 cm petri dish | ||
| 10 cm petri dishes | ||
| blades | - | - |
| FACS Aria II (5 lasers, 100 um nozzle) | BD Bioscience | SCR_018091 |
| Hemocytometer | NEUBAUER | HL-8100204 I |
| 0.2 mm syringe filters | Thermo Fisher Scientific | NC9103939 |
| 40 mm cell strainers | Corning | CLS431750 |
| 50 mL syringes | - | - |
| QuadroMACS™ Separator | Miltenyi Biotec | 130-090-976 |
| Esky with ice | - | - |
| 50 ml and 15 ml falcon canonical centrifuge tubes | Fisher Scientific | 14-959-53A 14-432-22 |
| Pasteur pipettes (5 and 10 ml) | - | - |
| Eppendorf tips (10, 20, 200 and 1000 µl) | - | - |
| 5 ml FACS tubes | - | - |
| Trypan Blue stain (0.4%) | Thermo Fisher Scientific | T10282 |
| 100% Ethanol | ||
| 70 ml specimen Jar | Sarstedt | SAR00002 |
| LS columns | Miltenyi Biotec | 130-042-401 |
| Water bath | ||
| Timer | ||
| Animals | ||
| C57BL/6J mice | The Jackson Laboratory | JAX:000664 |
| PdgfraGFP/+ mice | The Jackson Laboratory | MGI:2663656 |
Before you start:
· Set all centrifuges to 4 oC
· Set the water bath to 37 oC and adjust the water level, enough to cover half of the specimen jar.
· Place the collagenase type II stock bottle in a desiccator at room temperate (RT) for at least 30 mins before opening.
· Pre-warm the red cell lysis buffer to RT.
Reagents to prepare:
· FACS buffer: Prepare 2% FBS in PBS and filter through 0.2 µm syringe filter.
· Collagenase type II in PBS (working concentration: 265 U/ml): Prepare fresh solution each time. Dissolve collagenase in PBS and filter through 0.2 µm syringe filter. Note: check the dry concentration (U/mg) of the protein as specified by the manufacturer and prepare normalized working solution based on the unit concentration. Prepare 15 ml of collagenase working solution for each heart sample.
· NOTE: Perform all steps on ice unless otherwise indicated.
Method:
1. Euthanize mouse by cervical dislocation.
2. Excise the heart and place it into a 10 cm petri dish containing PBS on ice. Rinse out as much blood as possible. Remove atria and great vessels with fine surgical scissors.
3. Transfer the heart to a fresh 6 cm petri dish and mince the tissue into small pieces (<~1 mm3) using a scalpel blade. Cover the tissue in a small volume of collagenase solution (~500 µl) to keep them moist during mincing. Note that excessive mincing of the heart tissue may cause considerable cell death
4. Add 5 ml collagenase solution to the dish and transfer the minced tissue into a 70 ml specimen jar using a Pasteur pipette. Mix by gently pipetting up and down to break down the clumps.
5. Incubate the specimen jar at 37 oC in water bath with shaker for 10 mins, intermittently pipette the tissue gently up and down a few times to break down clumps while avoiding bubble formation.
6. After 10 mins, let the solution settle before transferring the supernatant (suspension cells)
to a 50 ml Falcon tube by filtering through a 40 μm sterile cell strainer to remove undigested tissue and debris.
7. Add fresh 5 ml collagenase solution to the remaining undigested tissue in the specimen jar, mix well by pipetting up and down, and repeat steps 5-6 for a further two times. Pool the cell suspension of each heart in same designated 50 ml falcon tube. Centrifuge the cell suspension at 300g for 5 mins, discard the supernatant.
8. Resuspend the pellet in 1 mL of red cell lysis buffer and incubate at RT for 1 min. Note: Warm the red cell lysis buffer to RT prior to use. Centrifuge the tube at 300g for 5 mins and discard the supernatant.
9. Gently resuspend the pellet in 1 ml of ice-cold 2% FBS/PBS and transfer the contents into 1.5 ml Eppendorf tubes. Remove the supernatant carefully by aspiration. Note: handle the tubes gently; try to avoid disturbing the cell pellets at all washing steps which will help to reduce loss of cells.
10. Centrifuge the tube at 300g for 5 mins after washing, discard supernatant and resuspend the pellet in 200 µl of Dead Cell Removal MicroBeads (Miltenyi Biotec), mix well and incubate for 15 min at RT. The beads are conjugated to Annexin V which binds to the dead cell population.
11. During the incubation: prepare 15 ml (per heart) of 1× binding buffer from 20× stock solution; place MACS LS columns in the magnetic field of a QuadroMACS™ Separator and precondition the column with 3 ml of 1× binding buffer. Discard the flow-through.
12. Load cell suspension to the column and slowly add the remaining 12 ml of 1× binding buffer to each column, collect all the flow-through in to a 15 ml tube, placed on ice. The flow-through contains unlabeled live cells.
13. Centrifuge the tubes at 300g for 5 mins. After discarding the supernatant, resuspend the pellet in 1 ml of ice-cold 2% FBS/PBS and transfer the content into a 1.5 ml Eppendorf tube.
14. Centrifuge at 300g for 5 mins, resuspend the pellet in 500 µl of ice-cold 2% PBS/FBS.
15. Transfer 100 µl of cell suspension to a 5 ml FACS tube and dilute with ice-cold 2% PBS/FBS, if necessary. Add DAPI (final: 100 ng/ml) to cell suspension and sort for live single cells (DAPI-) as indicated in Figure 1.
16. When working with PdgfraGFP/+ or similar GFP-lineage-tagged mice, you need use unstained cells isolated simultaneously from the heart of a wild type mouse of the same background strain to estaFigure 1: FACS gating strategy. A typical workflow of sequential gating for doublet exclusion is shown Total events (A) were gated and subjected to FSC-based (B) and SSC-based (C) single cell gating. Live TIP cells were then gated as DAPI-negative fraction (D). In (E) we show the example of gating for GFP-positive cells isolated from PdgfraGFP/+ mice.
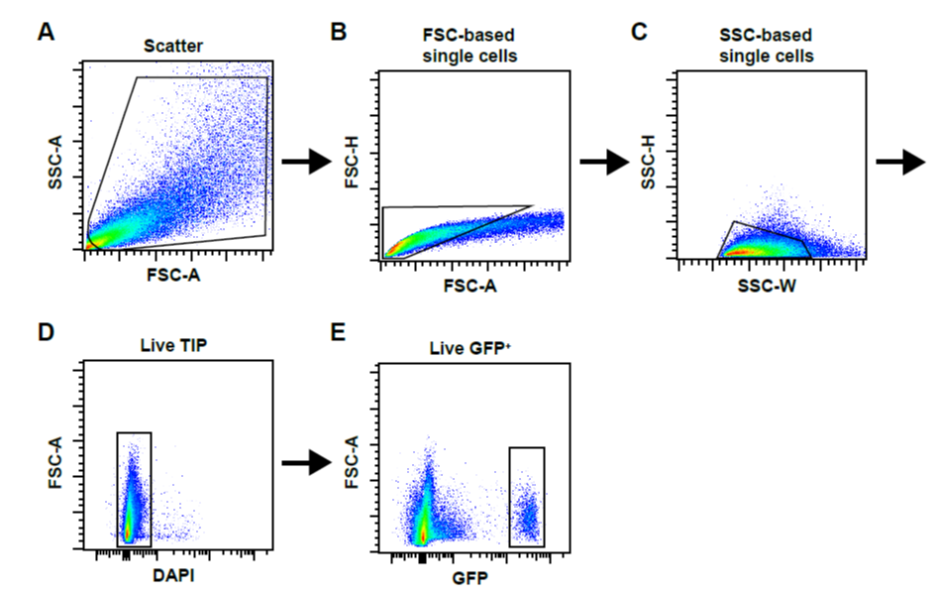
Figure 1: FACS gating strategy. A typical workflow of sequential gating for doublet exclusion is shown Total events (A) were gated and subjected to FSC-based (B) and SSC-based (C) single cell gating. Live TIP cells were then gated as DAPI-negative fraction (D). In (E) we show the example of gating for GFP-positive cells isolated from PdgfraGFP/+ mice.
Acknowledgements
This work was supported by funding from the National Health and Medical Research Council (1118576, 573707, 1105271, 1074286), Leducq Foundation (13CVD01, 15CVD03), Stem Cells Australia blished gates for GFP- cells.
- Farbehi, N, Janbandhu, V, Patrick, R, Xaymardan, M, Nordon, R R and Harvey, R P(2020). Protocols for FACS purification of cardiac total interstitial single cells from adult mouse cardiac ventricles. Bio-protocol Preprint. bio-protocol.org/prep615.
- Farbehi, N., Patrick, R., Dorison, A., Xaymardan, M., Janbandhu, V., Wystub-Lis, K., Ho, J. W., Nordon, R. E. and Harvey, R. P.(2019). Single-cell expression profiling reveals dynamic flux of cardiac stromal, vascular and immune cells in health and injury. eLife. DOI: 10.7554/eLife.43882
Category
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link


