Advanced Search
PCR-RFLP of G1 and G2 mutations in the human Apolipoprotein-L1 (APOL1) gene
Last updated date: Aug 25, 2020 Views: 1424 Forks: 0
PCR-RFLP of G1 and G2 mutations in the human Apolipoprotein-L1 (APOL1) gene
Abstract
Two Apolipoprotein-L1 (APOL1) renal risk variants, named G1 and G2, occur at high frequency in populations of recent African ancestry. Inheriting two risk variants of APOL1, in any combination, is associated with an increased risk of a number of non-diabetic kidney diseases, including focal segmental glomerulosclerosis (FSGS), hypertension-associated end-stage kidney disease, and HIV-associated nephropathy (Genovese et al. 2010; Kopp et al. 2011; Kasembeli et al. 2015). This recessive renal risk is balanced by a protective association against Human African trypanosomiasis, or sleeping sickness, a deadly disease caused by the protozoan parasite Trypanosoma brucei in sub-Saharan Africa. The G2 variant is associated with dominant protection from T.b. rhodesiense infection (the east African form of the disease), whereas carriage of at least one copy of the G1 variant is associated with reduced disease severity for the west African form of the disease, T.b. gambiense (Cooper et al. 2017).
G1 and G2 mutations are closely spaced in the final exon of APOL1 but located on separate haplotypes. G1 comprises two non-synonymous substitutions in near-perfect linkage disequilibrium.
· rs73885319 (c.1024A>G [p.S342G])
· rs60910145 (c.1152T>G [p.I384M])
G2 is found on an alternative haplotype and represents a two amino acid in-frame deletion
· rs71785313 (c.1164_1169del [p.N388_Y389del]).
All 3 mutations can be detected by PCR-RFLP.
Materials and reagents
1. DNAase-free ddH2O (e.g. Qiagen 129115)
2. Custom primers (e.g. Eurofins)
3. Standard PCR reagents (buffer, dNTPs, Taq polymerase [e.g. NEB M0267S])
4. Thin-walled PCR tubes
5. HindIII + buffer (NEB, R0104S)
6. NspI + buffer (NEB, R0602S)
7. MluCI + buffer (NEB, R0538S)
8. Agarose
9. Gel running buffer (e.g TAE or TBE)
10. Gel loading dye (e.g. NEB B7025S)
11. Ethidium bromide (e.g. Sigma Aldrich E1510)
12. 100bp DNA Ladder (e.g. NEB N3231S)
Equipment
1. Thermocycler
2. 37oC incubator or waterbath
3. DNA electrophoresis apparatus
4. Gel doc system
PCR protocol
Perform a 25uL PCR reaction to amplify a 458 bp product containing all three mutations from the final exon of APOL1.
Primers
Forward primer APOL1 F1 5’-AGACGAGCCAGAGCCAATCTTC-3’
Reverse primer APOL1 R2 5’-CACCATTGCACTCCAACTTGGC-3’
| PCR reaction mix (25uL) | ||
| Component | Volume (uL) | Final conc. |
| DNAase-free ddH2O | 16.25 | |
| Forward primer (10 μM) | 2.5 | 1μM |
| Reverse primer (10 μM) | 2.5 | 1μM |
| PCR mastermix (10x) | 2.5
| 45 mM Tris, 11 mM (NH4)2SO4, 4.5 mM MgCl2, 4.4 µM EDTA, 113 µg/ml BSA, 1mM dATP, 1mM dGTP, 1mM dTTP and 1mM dCTP |
| Taq polymerase (5 U/μl) | 0.25 | 1.25 U per reaction |
| DNA (10-200ng/μl) | 1 μl or 1mm FTA disc | |
Thermocycler program
| 1 cycle | 95 °C, 2 min |
| 32 cycles | 95 °C, 50 sec; 67°C, 50 sec; 70 °C, 1min |
| 1 cycle | 70 °C, 5 min |
Digest PCR product
Set up 3 independent digest reactions from the PCR product, one to test for each mutation.
G1 rs73885319 destroys a HindIII digest site
G1 rs60910145 creates a NspI digest site
G2 rs71785313 destroys a MluCI site
| Digest reaction mix (20uL) | |
| Component | Volume (uL) |
| DNAase-free ddH2O | 13.5 |
| 10x reaction buffer | 2.0 |
| Enzyme (5 U/μl to 10 U/μl) | 0.5 |
| PCR product | 4 |
2. Incubate at 37 °C for minimum 2 hr to O/N.
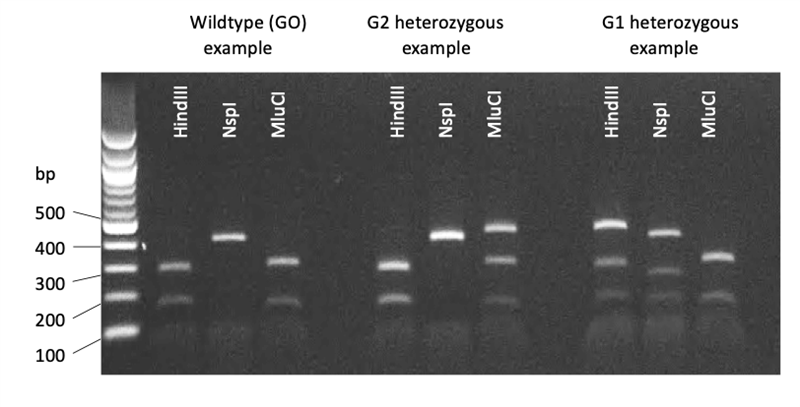
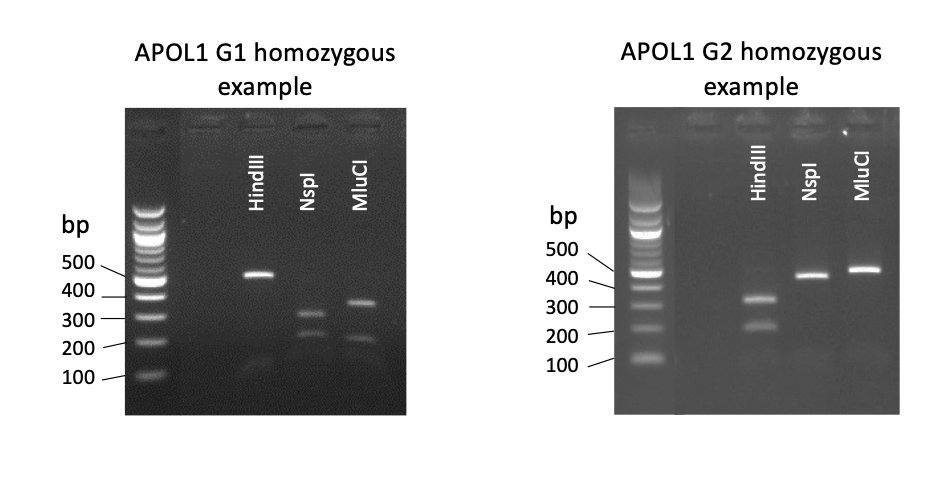
3. Resolve digest fragment sizes by gel electrophoresis of ~10uL of digest reaction + loading buffer on a 1.5-2% agarose gel, containing 0.1 ug/ml ethidium bromide and visualisation under UV light. Estimate fragment sizes by comparison to 100bp ladder.
| Expected PCR-RFLP produce sizes (bp) | |||
| Wildtype homozygous | Wildtype/mutant heterozygous | Mutant homozygous | |
| HindIII |
G1 rs73885319 destroys a HindIII digest site | ||
| 165 + 293 | 165 + 293 + 458 | 458 | |
| NspI | G1 rs60910145 creates a NspI digest site$ | ||
| 421 + 37 | 421 + 257 + 164 + 37 | 257 + 164 + 37 | |
| MluCI | G2 rs71785313 destroys a MluCI site | ||
| 305 + 153 | 305 + 153 + 458 | 458 | |
$Note there is another NspI site within the PCR product unaffected by the G1 or G2 mutations that produces a 37bp + 421bp digest fragments in all samples.
4. Data for each SNP (rs73885319, rs60910145, and rs71785313) is combined to generate the APOL1 genotype for each individual.
Representative example image
- Cooper, A(2020). PCR-RFLP of G1 and G2 mutations in the human Apolipoprotein-L1 (APOL1) gene. Bio-protocol Preprint. bio-protocol.org/prep465.
- Cooper, A., Ilboudo, H., Alibu, V. P., Ravel, S., Enyaru, J., Weir, W., Noyes, H., Capewell, P., Camara, M., Milet, J., Jamonneau, V., Camara, O., Matovu, E., Bucheton, B. and MacLeod, A.(2017). APOL1 renal risk variants have contrasting resistance and susceptibility associations with African trypanosomiasis. eLife. DOI: 10.7554/eLife.25461
Category
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link
