Advanced Search
Cell culture and treatment
Last updated date: Aug 17, 2020 Views: 953 Forks: 0
Cell culture and treatment of melanoma cells to assess PRMT5 effect on dsDNA/dsRNA-induced STING activation and type I IFN (IFNI) response
Procedure
- Murine melanoma cells (B16F10, YUMMER1.7) are cultured in complete DMEM supplemented with 10 % FBS and penicillin-streptomycin solution.
- Before stimulation, cells (5 ~ 7.5 X 105 cells in 2ml of complete DMEM per well) were plated into 6-well culture plate (Thermo Fisher Scientific) in 5 % CO2 and 37°C.
- Induce the loss- or gain-of-PRMT5 functions in cells by
- Stably expressing short-hairpin RNAs against PRMT5
- Treating cells with PRMT5 inhibitor (EPZ015666, 10 mM or GSK3326595, 10 mM) for 24 ~ 48 hrs
- Stably expressing PRMT5 and WDR77
- Plated cells are transfected with dsDNA (V70mer, 1.5 mg/ml for assessing signaling activation, 0.5 mg/ml for assessing transcriptional activation of IFNI genes) or dsRNA [poly(I:C), 250ng/ml] using jetPrime [1:2 ( mg of DNA: ml of jetPrime) (w/v) ratio].
- Transfected cells are incubated in 5 % CO2 and 37°C and harvested at 0, 3, 6 and 9 hrs for assessing activation of signaling cascade or 0 and 6 hrs for assessing transcriptional activation of IFNI genes.
- After stimulation, cells are washed three times with PBS and lysed with
- 150 ml/well of RIPA buffer by three cycles of freezing-thawing (SDS-PAGE or semi-Native PAGE) on dry ice
- 150 ml/well Native-lysis buffer by rotating for 30 min at 4 °C [BlueNative (BN) PAGE]
- 250ul of lysis buffer in GenEluteTM (Sigma, #RTN70) for qPCR analysis
- Lysates for protein analysis are collected and assessed protein concentration using Pierce Coomassie (Bradford) protein assay kit (#23200, ThermoFisher scientific).
- Lysates for RNA purification are further processed for purify total RNA using GenEluteTM and, then, subjected to cDNA synthesis using Applied BiosystemsTM High-capacity cDNA reverse transcription kit (ThermoFisher Scientific, #4363814).
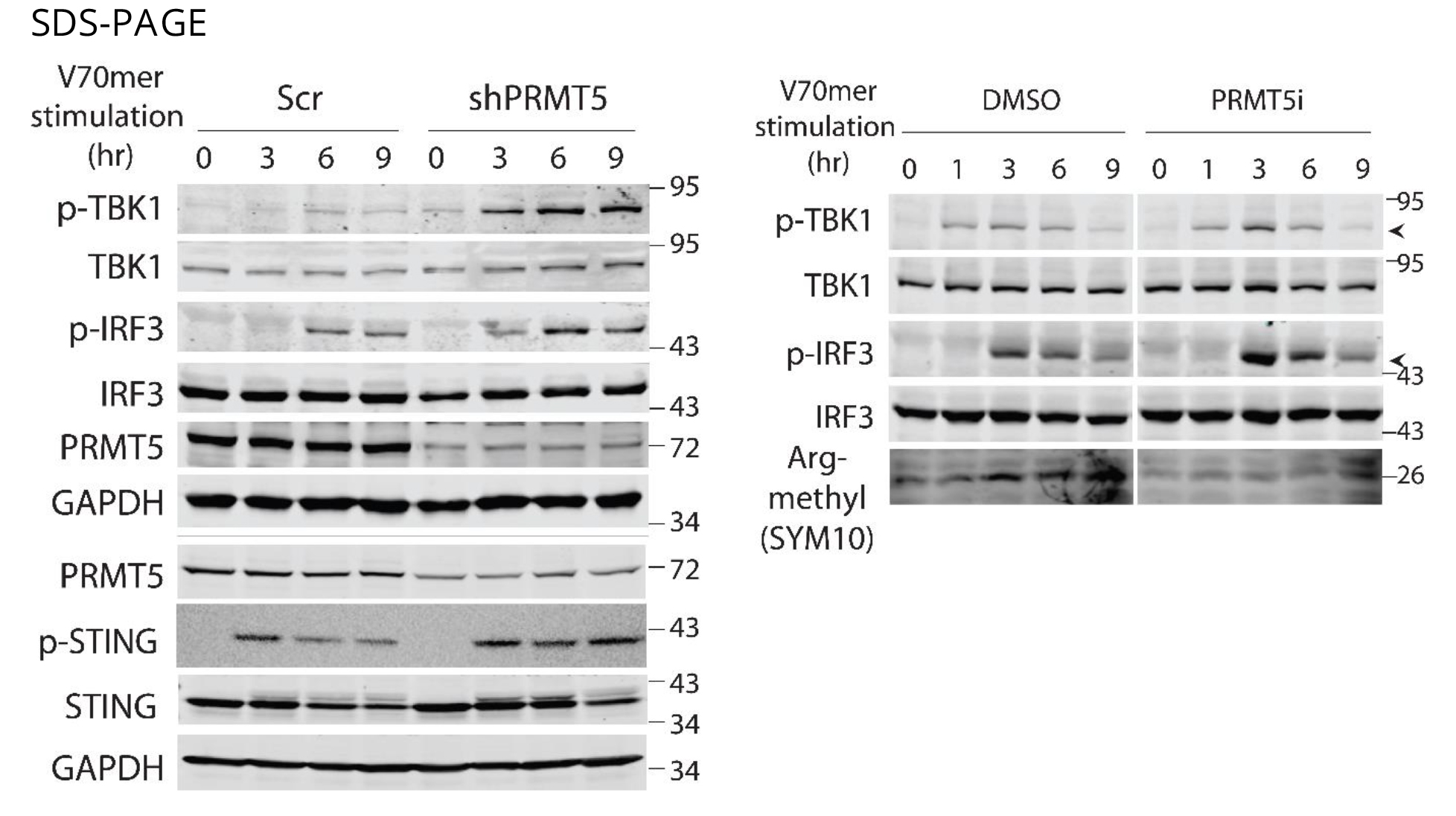
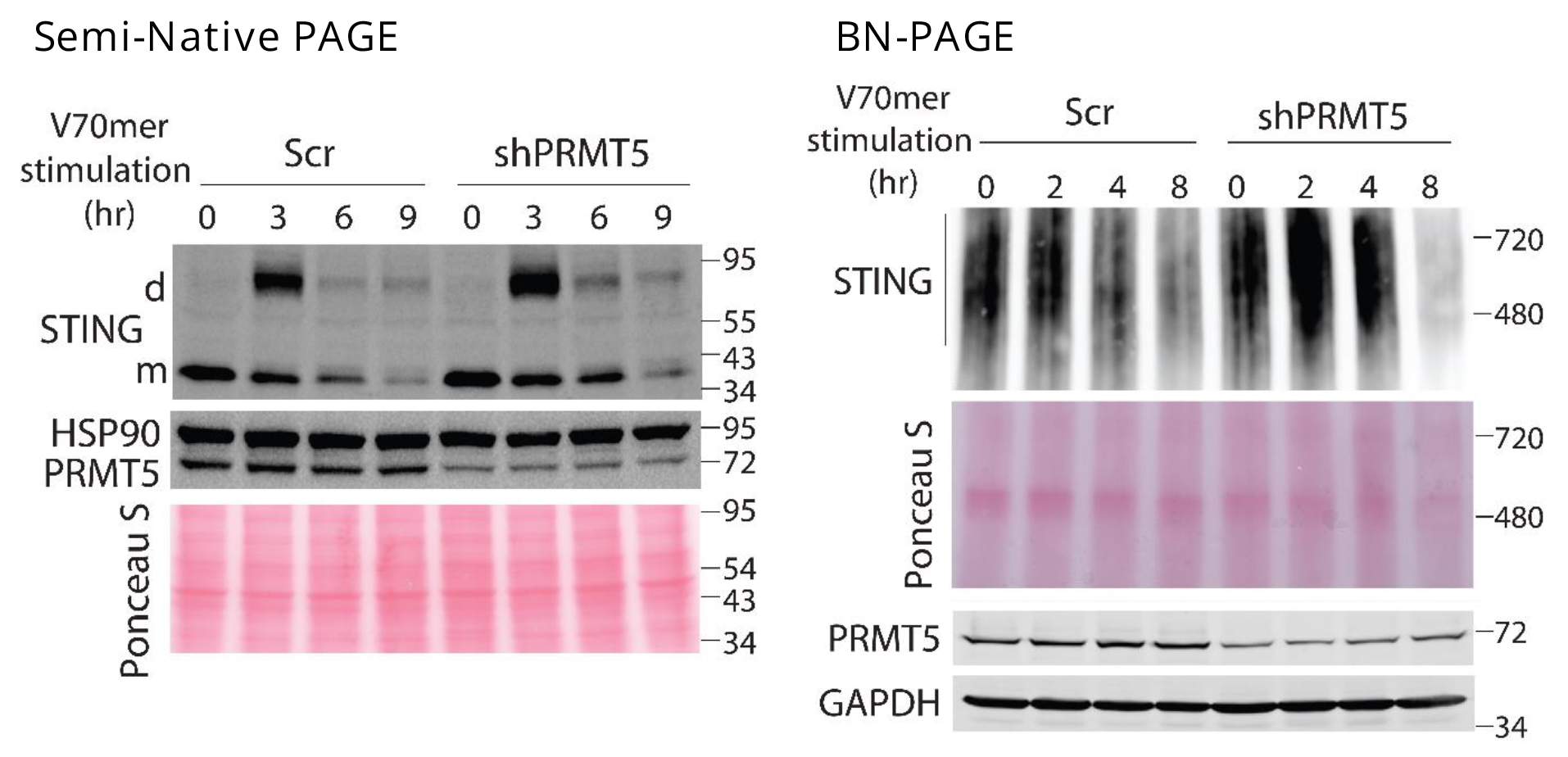
- The activation of signaling cascade by dsDNA stimulation in the prepared protein samples (40 ~ 60 mg/lane, need to adjust the amount of samples depending on the signal intensity of interesting protein) is assessed by SDS-PAGE, Semi-Native PAGE or BN PAGE and appropriate antibodies listed below
- The transcriptional activation of IFNI genes (IFNB1, CCL5 and CXCL10) is assessed by qPCR using primers listed below.
Reagents
- Short-hapirpin RNAs against PRMT5: shPRMT-1 (TRCN0000181891) and shPRMT5-2 (TRCN00001182569)
- EPZ015666 (10mM in DMSO, Apexbio, #B4989), GSK3326595 (10mM in DMSO, Chemitek, #CT-GSK332)
- pLentipuro-PRMT5; murine Prmt5 gene is cloned into pLenti-puro (Addgene #39481) and pLX304-WDR77; murine Wdr77 gene is cloned into pLX304 (Addgene #25890)
- jetPrime (Polyplus transfection, #114-15)
- dsDNA (V70mer): annealed forward (5'- CCATCAGAAAGAGGTTTAATATTTTTGTGAGACCATCGGGGCCGCGCCTCCCCCGCGAGGCCGCCGGCG-3′-3') and reverse (5'-CGCCGGCGGCCTCGCGGGGGAGGCGCGGCCCCGATGGTCTCACAAAAATATTAAACCTCTTTCTGATGG-3') strands
- Poly(I:C): Low molecular weight (LMW, #tlrl-picw) or High molecular weight (HMW, #tlrl-pic) from InvivoGen
- RIPA buffer: 50 mM Tris-HCl, pH7.4, 1 % (v/v) NP40, 0.1 % (w/v) sodium deoxycholate, 0.1 % (w/v) sodium dodecyl sulfate, 150 mM NaCl, 1 mM EDTA, and Pierce protease (#A32955)/phosphatase (#A32957) inhibitors tablets (ThermoFisher Scientific)
- Native lysis buffer: 20 mM HEPES, pH7.0, 25 mM NaCl, 10 % glycerol, 1% DDM (n-dodecyl b-D-maltoside, ThermoFisher Scientific #BN2005), Pierce protease inhibitor tablet
- Tested antibodies: PRMT5 (A-11, Santa Cruz Biotechnology), WDR77 (FG-4, Santa Cruz Biotechnology), GAPDH (C65, Santa Cruz Biotechnology), HSP90 (F-8, Santa Cruz Biotechnology), Symmetric-dimethyl-Arginine (SYM10, EMD Millipore or #13222, Cell Signaling), TBK1 (#3013, Cell Signaling), phospho-TBK1 (D52C2, Cell Signaling), IRF3 (D83B9, Cell Signaling), phospho-IRF3 (4D4G or D601M, Cell Signaling), STING (D2P2F or D1V5L, Cell Signaling), phospho-STING (D1C4T, Cell Signaling), STING for semi-Native PAGE and BN PAGE (D2P2F, Cell Signaling)
Tested primers:
H3.3 (used for an internal control for normalization):
5'-TGTGGCCCTCCGTGAAATC-3', 5'-GGCATAATTGTTACACGTTTGGC-3'
mouse CXCL10:
5'-CCAAGTGCTGCCGTCATTTTC-3', 5'-GGCTCGCAGGGATGATTTCAA-3'
mouse CCL5:
5'-CTCACCATATGGCTCGGACA-3', 5'-CTTCTCTGGGTTGGCACACA-3'
mouse IFNB1:
5'-CAGCTCCAAGAAAGGACGAAC-3', 5'-GGCAGTGTAACTCTTCTGCAT-3'
mouse PRMT5:
5'-CTGAATTGCGTCCCCGAAATA-3', 5'-AGGTTCCTGAATGAACTCCCT-3'
mouse WDR77:
5'-CTTGCTGTGCTGGATTCAAGC-3', 5'-CAACTGTGGTAAGAAGGGAGTG-3'
How to cite: Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
1. Kim, H., and Ronai, Z.A. (2020). Cell culture and treatment of melanoma cells to assess PRMT5 effect on DNA/RNA-induced STING activation and type I IFN (IFNI) response. Bio-protocol.
2. Kim, H., Kim, H., Feng, Y., Li, Y., Tamiya, H., Tocci, S., and Ronai, Z.A. (2020). PRMT5 control of cGAS/STING and NLRC5 pathways defines melanoma response to antitumor immunity. Sci Transl Med. DOI: 10.1126/scitranslmed.aaz5683
Copyright: Content may be subjected to copyright.
- Kim, H and Ronai, Z(2020). Cell culture and treatment. Bio-protocol Preprint. bio-protocol.org/prep451.
- Kim, H., Kim, H., Feng, Y., Li, Y., Tamiya, H., Tocci, S. and Ronai, Z. A.(2020). PRMT5 control of cGAS/STING and NLRC5 pathways defines melanoma response to antitumor immunity . Science Translational Medicine 12(551). DOI: 10.1126/scitranslmed.aaz5683
Category
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Tips for asking effective questions
+ Description
Write a detailed description. Include all information that will help others answer your question including experimental processes, conditions, and relevant images.
Share
Bluesky
X
Copy link
