Advanced Search
Yeast Three-Hybrid Screening and Validation Experiment Protocol
Last updated date: Oct 21, 2024 Views: 169 Forks: 1
Instruments:
PCR Machine**: Used for PCR amplification.
Temperature-Controlled Centrifuge**: Used for centrifugal separation.
Thermostatic Water Bath**: Used for yeast transformation steps.
Gel Imaging System**: Used to observe the results of agarose gel electrophoresis.
Agarose Gel Electrophoresis Equipment**: Used for the separation and purification of DNA fragments.
Micropipettes**: Used for precise reagent dispensing.
Temperature-Controlled Incubator**: Used for the cultivation of bacterial and yeast strains.
Laminar Flow Cabinet (Sterile Workstation)**: Used for sterile operations.
Reagents and Chemicals:
pBRIDGE Vector**: Used for constructing the bait vector.
pGADT7 Vector**: Used for constructing the prey vector.
KOD FX DNA Polymerase**: Used for PCR amplification.
dNTPs (Deoxyribonucleotide Triphosphates)**: Substrates for PCR reactions.
Restriction Enzymes (NotI-HF, BglII-HF, EcoRI-HF, SalI-HF, BamHI-HF)**: Used for cutting vectors and insert fragments.
rCutSmart Buffer**: Used for restriction enzyme reactions.
ClonExpress® Ultra One Step Cloning Kit**: Used for homologous recombination cloning.
EasyPure Quick Gel Extraction Kit**: Used for extracting DNA fragments from agarose gels.
Yeast Competent Cells (Y2HGold, Strain cc309)**: Used for transforming constructed vectors.
PEG/LiAc Solution**: Used for yeast transformation.
Carrier DNA**: Used as auxiliary DNA during yeast transformation.
SD Medium (Synthetic Defined Medium)**: Synthetic dropout medium for selection and screening of yeast strains.
X-α-Gal and 3-AT**: Used for yeast colony screening and auto-activation testing.
Antibiotics (e.g., Ampicillin)**: Used for bacterial culture and plasmid selection.
Plasmid Extraction Kit**: Used for extracting plasmid DNA from bacteria.
T7 and 3’AD Sequencing Primers**: Used for DNA sequencing.
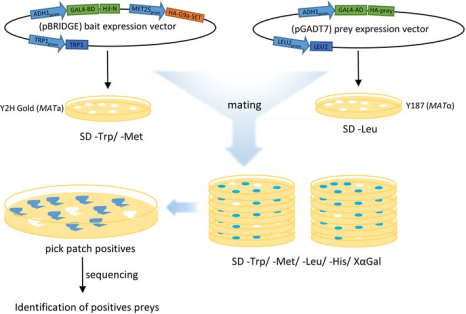
Schematic workflow of the Y3H experiment
Vectors used in this study were part of the Matchmaker Gold Yeast Two-Hybrid System (Takara Bio, Mountain View, CA, USA). The pBRIDGE vector was used as a backbone for the bait construct. To design a bait that will be recognized in a methylation-dependent manner, the catalytic SET domain of the G9a PKMT (G9a-SET; residues 931–1210) together with a nuclear localization sequence (NLS) and an HA-tag was cloned under the MET25 promoter in the MCS II region of the pBRIDGE vector using NotI and BglII restriction sites. On the same vector, the N-terminal twenty residues of histone H3 (referred later as an H3N) were cloned in fusion with the GAL4-binding domain (GAL4-BD) under an ADH1 promoter in the MCS I using EcoRI and BamHI restriction sites. The bait plasmid with H3N located C-terminally with respect to GAL4-BD was also cloned. The pBRIDGE vector containing GAL4-BD-H3N but without G9a-SET was also generated as a negative control. The preys were expressed in the pGADT7 vector(Rawłuszko-Wieczorek AA, Knodel F, Tamas R, Dhayalan A, Jeltsch A. Identification of protein lysine methylation readers with a yeast three-hybrid approach. Epigenetics Chromatin. 2018 Jan 25;11(1):4.)
The purpose of the yeast three-hybrid (Y3H) experiment is to verify the effect of gene X on the interaction between genes A and B, given their known relationship. Common methods include constructing gene A into the pGADT7 vector, and genes B and X into the MCSI and MCSII sites of the pBridge vector, respectively, and verifying the interaction through co-transformation into Y2HGold yeast cells.
1. Construction of pBridge-B-X Vector
1.1 **PCR Amplification of Bait-B with Adapter Sequences**
- Bait sequence synthesis rules:
- B-F: `tgactgtatcgccgg` + Bait CDS first 20bp
- B-R: `tagcttggctgcagg` + Bait CDS last 20bp (reverse complement)
1.2 **PCR Amplification System for Bait Gene B with Adapters (using KOD FX, TOYOBO KFX-101)**
- Before preparing the reaction mixture, mix all reagents except KOD FX (enzyme solution) thoroughly. Thaw frozen reagents on ice before use.
- 2x PCR buffer: 25 μl
- 2mM dNTPs: 10 μl
- B-F Primer: 1.5 μl
- B-R Primer: 1.5 μl
- Plant cDNA: 0.2 μg
- KOD FX (1.0U/μl): 1 μl
- ddH2O: up to 50 μl
- Add KOD FX (enzyme solution) last, mix the reaction mixture thoroughly using a vortex, spin down, and then perform PCR.
1.3 **PCR Amplification of Bait-X with Adapter Sequences**
- Bait sequence synthesis rules:
- X-F: `gaagagaaaggtggc` + Bait CDS first 20bp
- X-R: `ggagatcagcccgaa` + Bait CDS last 20bp (reverse complement)
1.4 **PCR Amplification System for Bait Gene X with Adapters (using KOD FX, TOYOBO KFX-101)**
- Before preparing the reaction mixture, mix all reagents except KOD FX (enzyme solution) thoroughly. Thaw frozen reagents on ice before use.
- 2x PCR buffer: 25 μl
- 2mM dNTPs: 10 μl
- X-F Primer: 1.5 μl
- X-R Primer: 1.5 μl
- Plant cDNA: 0.2 μg
- KOD FX (1.0U/μl): 1 μl
- ddH2O: up to 50 μl
- Add KOD FX (enzyme solution) last, mix the reaction mixture thoroughly using a vortex, spin down, and then perform PCR.
1.5 **Linearization of pGBKT7 Plasmid**
- Remove pGBKT7 plasmid from -20°C freezer, place 10x rCutSmart buffer on ice until completely dissolved, and prepare the following system:
- 10x rCutSmart buffer: 5 μl
- EcoRI-HF: 1 μl
- SalI-HF: 1 μl
- pBridge Plasmid DNA: 1 μg
- ddH2O: up to 50 μl
- Use a temperature-controlled PCR machine with the following program:
- 37°C for 45 min
- 65°C for 45 sec
- After the reaction, transfer to a 4°C refrigerator for storage.
1.6 **Agarose Gel Electrophoresis and Gel Extraction (EasyPure Quick Gel Extraction Kit, TransGen Biotech, EG101)**
- Method reference: Wang, S., et al. (2024). Application of the Nicotiana Allergic Necrosis Assay for the Validation of Protein-Protein Interactions between Fungal Effectors and Plant Receptor Kinases. Bio-protocol Preprint. bio-protocol.org/prep2729.
1.7 **Homologous Recombination to Construct pBridge-B Vector (ClonExpress® Ultra One Step Cloning Kit, Vazyme, C115)**
- Calculate the dosage of linearized vector and insert fragment:
- Optimal cloning vector dosage = [0.02 × vector base pairs] ng (0.03 pmol)
- Optimal insert fragment dosage = [0.04 × insert fragment base pairs] ng (0.06 pmol)
- Note: To ensure accurate sample addition, dilute the linearized vector and insert fragment appropriately before preparing the recombination reaction system, with each component added in a volume not less than 1 μl.
- Prepare the following reaction system on ice:
- Linearized vector pBridge: X μl
- Insert fragment Bait: B μl
- 2× ClonExpress Mix: 5 μl
- ddH2O: to 10 μl
- Mix gently using a pipette (do not vortex), spin down briefly to collect the reaction mixture at the bottom of the tube.
- Use a temperature-controlled PCR machine with the following program:
- 50°C for 30 min
- After the reaction, transfer to a 4°C refrigerator for storage.
1.8 **Sequencing of pBridge-B**
- Sequencing primers and sequences:
- 5' sequencing primer: 5'-TCATCGGAAGAGAGTAGT-3'
- 3' sequencing primer: 5'-AATTTATTTCTTTTCGGATAA-3'
- After sequencing, expand the E. coli culture and extract the plasmid. Method reference: Wang, S., et al. (2024). Application of the Nicotiana Allergic Necrosis Assay for the Validation of Protein-Protein Interactions between Fungal Effectors and Plant Receptor Kinases. Bio-protocol Preprint. bio-protocol.org/prep2729.
1.9 **Linearization of pBridge-B Plasmid**
- Remove pBridge-B plasmid from -20°C freezer, place 10x rCutSmart buffer on ice until completely dissolved, and prepare the following system:
- 10x rCutSmart buffer: 5 μl
- NotI-HF: 1 μl
- BglII-HF: 1 μl
- pBridge-B Plasmid DNA: 1 μg
- ddH2O: up to 50 μl
- Use a temperature-controlled PCR machine with the following program:
- 37°C for 45 min
- 65°C for 45 sec
- After the reaction, transfer to a 4°C refrigerator for storage.
1.10 **Agarose Gel Electrophoresis and Gel Extraction (EasyPure Quick Gel Extraction Kit, TransGen Biotech, EG101)**
- Method reference: Wang, S., et al. (2024). Application of the Nicotiana Allergic Necrosis Assay for the Validation of Protein-Protein Interactions between Fungal Effectors and Plant Receptor Kinases. Bio-protocol Preprint. bio-protocol.org/prep2729.
1.11 **Homologous Recombination to Construct pBridge-B-X Vector (ClonExpress® Ultra One Step Cloning Kit, Vazyme, C115)**
- Calculate the dosage of linearized vector and insert fragment:
- Optimal cloning vector dosage = [0.02 × vector base pairs] ng (0.03 pmol)
- Optimal insert fragment dosage = [0.04 × insert fragment base pairs] ng (0.06 pmol)
- Note: To ensure accurate sample addition, dilute the linearized vector and insert fragment appropriately before preparing the recombination reaction system, with each component added in a volume not less than 1 μl.
- Prepare the following reaction system on ice:
- Linearized vector pBridge-B: X μl
- Insert fragment Bait: X μl
- 2× ClonExpress Mix: 5 μl
- ddH2O: to 10 μl
- Mix gently using a pipette (do not vortex), spin down briefly to collect the reaction mixture at the bottom of the tube.
- Use a temperature-controlled PCR machine with the following program:
- 50°C for 30 min
- After the reaction, transfer to a 4°C refrigerator for storage.
1.12 **Sequencing of pBridge-B-X**
- Sequencing primers and sequences:
- 5' sequencing primer: 5'-TCATCGGAAGAGAGTAGT-3'
- 3' sequencing primer: 5'-AATTTATTTCTTTTCGGATAA-3'
- After sequencing, expand the E. coli culture and extract the plasmid. Method reference: Wang, S., et al. (2024). Application of the Nicotiana Allergic Necrosis Assay for the Validation of Protein-Protein Interactions between Fungal Effectors and Plant Receptor Kinases. Bio-protocol Preprint. bio-protocol.org/prep2729.
#### 2. Construction of pGADT7-A Vector
2.1 **PCR Amplification of Prey with Adapter Sequences**
- Prey sequence synthesis rules:
- A-F: `GCCATGGAGGCCAGTGAA` + Prey CDS first 20bp
- A-R: `AGCTCGAGCTCGATGGAT` + Prey CDS last 20bp (reverse complement)
2.2 **PCR Amplification System for Prey Gene X with Adapters (using KOD FX, TOYOBO KFX-101)**
- Before preparing the reaction mixture, mix all reagents except KOD FX (enzyme solution) thoroughly. Thaw frozen reagents on ice before use.
- 2x PCR buffer: 25 μl
- 2mM dNTPs: 10 μl
- A-F Primer: 1.5 μl
- A-R Primer: 1.5 μl
- Prey cDNA: 0.2 μg
- KOD FX (1.0U/μl): 1 μl
- ddH2O: up to 50 μl
- Add KOD FX (enzyme solution) last, mix the reaction mixture thoroughly using a vortex, spin down, and then perform PCR.
2.3 **PCR Amplification of Prey Gene X with Adapters**
- Use a temperature-controlled PCR machine with the following program:
- Predenature at 94°C for 2 min.
- Denature at 98°C for 10 sec.
- Annealing at (Tm-5)°C for 30 sec.
- Extension at 68°C for 1kb/min.
- Set 33 cycles for Denature to Extension.
- Final extension at 68°C for 7 min.
- After the reaction, transfer to a 4°C refrigerator for storage.
2.4 **Linearization of pGADT7 Plasmid**
- Remove pGADT7 plasmid from -20°C freezer, place 10x rCutSmart buffer on ice until completely dissolved, and prepare the following system:
- 10x rCutSmart buffer: 5 μl
- EcoR I-HF: 1 μl
- BamH I-HF: 1 μl
- pGADT7 Plasmid DNA: 1 μg
- ddH2O: up to 50 μl
- Use a temperature-controlled PCR machine with the following program:
- 37°C for 45 min
- 65°C for 45 sec
- After the reaction, transfer to a 4°C refrigerator for storage.
2.5 **Agarose Gel Electrophoresis and Gel Extraction (EasyPure Quick Gel Extraction Kit, TransGen Biotech, EG101)**
- Method reference: Wang, S., et al. (2024). Application of the Nicotiana Allergic Necrosis Assay for the Validation of Protein-Protein Interactions between Fungal Effectors and Plant Receptor Kinases. Bio-protocol Preprint. bio-protocol.org/prep2729.
2.6 **Homologous Recombination to Construct Vector pGADT7-A (ClonExpress® Ultra One Step Cloning Kit, Vazyme, C115)**
- Calculate the dosage of linearized vector and insert fragment:
- Optimal cloning vector dosage = [0.02 × vector base pairs] ng (0.03 pmol)
- Optimal insert fragment dosage = [0.04 × insert fragment base pairs] ng (0.06 pmol)
- Note: To ensure accurate sample addition, dilute the linearized vector and insert fragment appropriately before preparing the recombination reaction system, with each component added in a volume not less than 1 μl.
- Prepare the following reaction system on ice:
- Linearized vector pGADT7: X μl
- Insert fragment Bait: A μl
- 2× ClonExpress Mix: 5 μl
- ddH2O: to 10 μl
- Mix gently using a pipette (do not vortex), spin down briefly to collect the reaction mixture at the bottom of the tube.
- Use a temperature-controlled PCR machine with the following program:
- 50°C for 30 min
- After the reaction, transfer to a 4°C refrigerator for storage.
2.7 **Sequencing of pGADT7-A**
- Sequencing primers and sequences:
- T7 sequencing primer
- 3’AD sequencing primer
- After sequencing, expand the E. coli culture and extract the plasmid. Method reference: Wang, S., et al. (2024). Application of the Nicotiana Allergic Necrosis Assay for the Validation of Protein-Protein Interactions between Fungal Effectors and Plant Receptor Kinases. Bio-protocol Preprint. bio-protocol.org/prep2729.
#### 3. Auto-Activation Test of pGBKT7-B-X
3.1 **Transformation of Y2HGold Competent Cells**
- Take 100 µl of Y2HGold competent cells (Coolaber: cc309) thawed on ice, add 1 µg each of pre-cooled pGADT7 + pBridge-B-X and pGADT7, 10 µl of Carrier DNA (95-100°C for 5 min, quick chill in ice water, repeat once), and 500 µl of PEG/LiAc (333 µL of 50% PEG3350 solution, 83.5 µL of 1M LiAc solution, 83.5 µL of 10x TE buffer), mix by pipetting several times, incubate at 30°C for 30 min (invert 6-8 times at 15 min).
- Place the tube in a 42°C water bath for 15 min (invert 6-8 times at 7.5 min).
- Centrifuge at 5000 rpm for 40 s, discard the supernatant, resuspend in 400 µl of ddH2O, centrifuge for 30 s, discard the supernatant.
- Resuspend in 150 µl of sterile water, then take 50 µL each to spread on SD/-Trp/-Met, SD/-Trp/-Met/X-α-Gal, and SD/-Trp/-Met/X-α-Gal/3-AT solid selection media, incubate at 29°C for 48-96h; on different concentrations of 3-AT (10 mM, 20 mM, 30 mM, 40 mM, 50 mM, 60 mM, 70 mM, 80 mM) plates, the fewest yeast colonies (and not blue) or none at all indicate the best 3-AT concentration (best inhibitory concentration, minimum inhibitory concentration, background expression concentration, auto-activation concentration).
#### 4. Yeast Three-Hybrid Dot-Blot Verification
4.1 **Transformation of Y2HGold Competent Cells**
- Take 100 µl of Y2HGold competent cells (Coolaber: cc309) thawed on ice, add 1 µg each of negative control: pGADT7 and pBridge-B-X, experimental group: pGADT7-A and pBridge-B-X, and 10 µl of Carrier DNA (95-100°C for 5 min, quick chill in ice water, repeat once), 500 µl of PEG/LiAc, mix by pipetting several times, incubate at 30°C for 30 min (invert 6-8 times at 15 min).
- Place the tube in a 42°C water bath for 15 min (invert 6-8 times at 7.5 min).
- Centrifuge at 5000 rpm for 40 s, discard the supernatant, resuspend in 400 µl of ddH2O, centrifuge for 30 s, discard the supernatant.
- Resuspend in 100 µl of sterile water, then take 50 µL each to spread on SD/-Leu/-Trp/-Met plates and SD/-Ade/-His/-Leu/-Trp/-Met/X-α-Gal/3-AT dropout plates (3-AT concentration as selected in auto-activation). Incubate at 30°C for 3 days, then observe colony growth.
- Note: Dilutions are 1:10, 1:100, 1:1000, 1:10,000, 1:100,000.
Supplement: Three-hybrid interaction verification dot-blotting (bait without auto-activation)
Y2HGold[pGADT7+pBridge-B-X] is spotted on SD/-Leu/-Trp (control group, can grow), SD/-Leu/-Trp/-His (experimental group), SD/-Leu/-Trp/-His/-Met (experimental group).
Three-hybrid interaction verification dot-blotting (bait with auto-activation)
Y2HGold[pGADT7+pBridge-B-X] is spotted on SD/-Leu/-Trp/-His/3-AT (control group), SD/-Leu/-Trp/-His/-Met/3-AT (control group); Y2HGold[pGADT7-A+pBridge-B-X] is spotted on SD/-Leu/-Trp/-His/3-AT (experimental group), SD/-Leu/-Trp/-His/-Met/3-AT (experimental group).
#### 5. Three-Hybrid Interaction Result Analysis
(Refer to the Y2HGold-pBridge System Yeast Three-Hybrid Interaction Verification Kit, Coolaber: YH3001-10T for discussion of results)
This is applicable only when A and B interact or do not interact, and the interaction relationship is known. The above experiments have determined whether the bait yeast strain has auto-activation, so the inhibition or promotion of A and B interaction by X can be analyzed in the presence or absence of auto-activation.
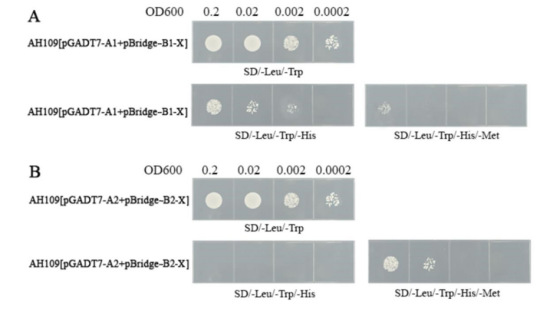
5.1 No Auto-Activation
The above experiments have determined that the bait has no auto-activation, so Y2HGold[pGADT7+pBridge-B-X] can be omitted as a control, and plates containing 3-AT can also be omitted. SD/-Leu/-Trp plates are used only as a control when X promotes the interaction between A and B (since no colonies are present on SD/-Leu/-Trp/-His plates). If the growth of Y2HGold[pGADT7-A+pBridge-B-X] on SD/-Leu/-Trp/-His and SD/-Leu/-Trp/-His/-Met plates is the same, then X has no effect on the relationship between A and B.
5.1.1 X Inhibits Interaction Between A and B
It is known from previous studies that A and B interact, and the bait has no auto-activation. Figure 1A, Y2HGold[pGADT7-A1+pBridge-B1-X] grows normally on SD/-Leu/-Trp plates (control), and the growth on SD/-Leu/-Trp/-His/-Met plates is weaker than on SD/-Leu/-Trp/-His plates (A1 and B1 interact), indicating that X inhibits the interaction between A1 and B1.
5.1.2 X Promotes Interaction Between A and B
It is known from previous studies that A and B do not interact, and the bait has no auto-activation. Figure 1B, Y2HGold[pGADT7-A2+pBridge-B2-X] grows normally on SD/-Leu/-Trp plates (control), does not grow on SD/-Leu/-Trp/-His plates (i.e., A2 and B2 do not interact), but can grow on SD/-Leu/-Trp/-His/-Met plates, indicating that X promotes the interaction between A2 and B2.
Three-hybrid interaction verification schematic (bait without auto-activation)
5.2 With Auto-Activation
The above experiments have determined that the bait has auto-activation, so plates containing 3-AT must be used to inhibit auto-activation of the bait and interaction between A and B. If the growth of Y2HGold[pGADT7-A+pBridge-B-X] on SD/-Leu/-Trp/-His and SD/-Leu/-Trp/-His/-Met plates is the same, then X has no effect on the relationship between A and B.
5.2.1 X Inhibits Interaction Between A and B
It is known from previous studies that A and B interact, and the bait has auto-activation. The concentration of 3-AT* on both plates is the same and can just inhibit the interaction of the double hybrid yeast (OD600=0.0002). Y2HGold[pGADT7-A3+pBridge-B3-X] grows weaker on SD/-Leu/-Trp/-His/-Met plates than on SD/-Leu/-Trp/-His plates (i.e., double hybrid yeast), and stronger than Y2HGold[pGADT7+pBridge-B3-X] (i.e., bait yeast), indicating that X inhibits the interaction between A3 and C3.
5.2.2 X Promotes Interaction Between A and B
It is known from previous studies that A and B do not interact, and the bait has auto-activation. Figure 2B, the concentration of 3-AT** on both plates is the same and can just inhibit the auto-activation of the bait yeast (OD600=0.0002). Y2HGold[pGADT7-A4+pBridge-B4-X] grows stronger on SD/-Leu/-Trp/-His/-Met plates than on SD/-Leu/-Trp/-His plates, and stronger than Y2HGold[pGADT7+pBridge-B4-X] (i.e., bait yeast), indicating that X promotes the interaction between A4 and B4.
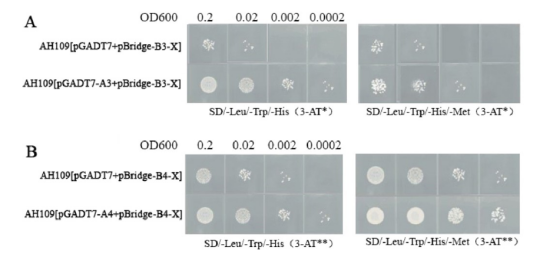
Three-hybrid interaction verification schematic (bait with auto-activation).
Reference
Maruta N, Trusov Y, Botella JR. Yeast Three-Hybrid System for the Detection of Protein-Protein Interactions. In: Botella J, Botella M, eds. Plant Signal Transduction. Methods in Molecular Biology. Vol 1363. Humana Press, New York, NY; 2016. https://doi.org/10.1007/978-1-4939-3115-6_12
Cottier S, Mönig T, Wang Z, Svoboda J, Boland W, Kaiser M, Kombrink E. The yeast three-hybrid system as an experimental platform to identify proteins interacting with small signaling molecules in plant cells: potential and limitations. Front Plant Sci. 2011;2:101. doi: 10.3389/fpls.2011.00101
Glass F, et al. The Yeast Three-Hybrid System for Protein Interactions. Methods Mol Biol. 2018;1794:61-73. doi: 10.1007/978-1-4939-7871-7_5
Tirode F, Malaguti C, Romero F, Attar R, Camonis J, Egly JM. A conditionally expressed third partner stabilizes or prevents the formation of a transcriptional activator in a three-hybrid system. J Biol Chem. 1997;272:22995–22999.
SenGupta DJ, Zhang B, Kraemer B, Pochart P, Fields S, Wickens M. A three-hybrid system to detect RNA–protein interactions in vivo. Proc Natl Acad Sci. 1996;93:8496–8501.
Kraemer B, Zhang B, SenGupta D, Fields S, Wickens M. Using the yeast three-hybrid system to detect and analyze RNA–protein interactions. Methods Enzymol. 2000;328:297–321.
Appendix: Vectors used in yeast one hybrid(Y3H) experiments
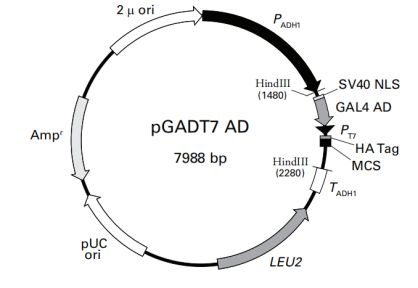

1.Vector Name: pGADT7
Origin of Replication: pUC
Promoter: ADH1
Vector Size: 7987 bp
5' Sequencing Primer and Sequence: 5'-CTATTCGATGATGAAGATACCCC-3'
3' Sequencing Primer and Sequence: 5'-GTGAACTTGCGGGGTTTTTCAG-3'
Vector Resistance: Ampicillin
Selection Marker: LEU2
>pGADT7 vector sequence
TGCATGCCTGCAGGTCGAGATCCGGGATCGAAGAAATGATGGTAAATGAAATAGGAAATCAAGGAGCATGAAGGCAAAAGACAAATATAAGGGTCGAACGAAAAATAAAGTGAAAAGTGTTGATATGATGTATTTGGCTTTGCGGCGCCGAAAAAACGAGTTTACGCAATTGCACAATCATGCTGACTCTGTGGCGGACCCGCGCTCTTGCCGGCCCGGCGATAACGCTGGGCGTGAGGCTGTGCCCGGCGGAGTTTTTTGCGCCTGCATTTTCCAAGGTTTACCCTGCGCTAAGGGGCGAGATTGGAGAAGCAATAAGAATGCCGGTTGGGGTTGCGATGATGACGACCACGACAACTGGTGTCATTATTTAAGTTGCCGAAAGAACCTGAGTGCATTTGCAACATGAGTATACTAGAAGAATGAGCCAAGACTTGCGAGACGCGAGTTTGCCGGTGGTGCGAACAATAGAGCGACCATGACCTTGAAGGTGAGACGCGCATAACCGCTAGAGTACTTTGAAGAGGAAACAGCAATAGGGTTGCTACCAGTATAAATAGACAGGTACATACAACACTGGAAATGGTTGTCTGTTTGAGTACGCTTTCAATTCATTTGGGTGTGCACTTTATTATGTTACAATATGGAAGGGAACTTTACACTTCTCCTATGCACATATATTAATTAAAGTCCAATGCTAGTAGAGAAGGGGGGTAACACCCCTCCGCGCTCTTTTCCGATTTTTTTCTAAACCGTGGAATATTTCGGATATCCTTTTGTTGTTTCCGGGTGTACAATATGGACTTCCTCTTTTCTGGCAACCAAACCCATACATCGGGATTCCTATAATACCTTCGTTGGTCTCCCTAACATGTAGGTGGCGGAGGGGAGATATACAATAGAACAGATACCAGACAAGACATAATGGGCTAAACAAGACTACACCAATTACACTGCCTCATTGATGGTGGTACATAACGAACTAATACTGTAGCCCTAGACTTGATAGCCATCATCATATCGAAGTTTCACTACCCTTTTTCCATTTGCCATCTATTGAAGTAATAATAGGCGCATGCAACTTCTTTTCTTTTTTTTTCTTTTCTCTCTCCCCCGTTGTTGTCTCACCATATCCGCAATGACAAAAAAATGATGGAAGACACTAAAGGAAAAAATTAACGACAAAGACAGCACCAACAGATGTCGTTGTTCCAGAGCTGATGAGGGGTATCTCGAAGCACACGAAACTTTTTCCTTCCTTCATTCACGCACACTACTCTCTAATGAGCAACGGTATACGGCCTTCCTTCCAGTTACTTGAATTTGAAATAAAAAAAAGTTTGCTGTCTTGCTATCAAGTATAAATAGACCTGCAATTATTAATCTTTTGTTTCCTCGTCATTGTTCTCGTTCCCTTTCTTCCTTGTTTCTTTTTCTGCACAATATTTCAAGCTATACCAAGCATACAATCAACTCCAAGCTTTGCAAAGATGGATAAAGCGGAATTAATTCCCGAGCCTCCAAAAAAGAAGAGAAAGGTCGAATTGGGTACCGCCGCCAATTTTAATCAAAGTGGGAATATTGCTGATAGCTCATTGTCCTTCACTTTCACTAACAGTAGCAACGGTCCGAACCTCATAACAACTCAAACAAATTCTCAAGCGCTTTCACAACCAATTGCCTCCTCTAACGTTCATGATAACTTCATGAATAATGAAATCACGGCTAGTAAAATTGATGATGGTAATAATTCAAAACCACTGTCACCTGGTTGGACGGACCAAACTGCGTATAACGCGTTTGGAATCACTACAGGGATGTTTAATACCACTACAATGGATGATGTATATAACTATCTATTCGATGATGAAGATACCCCACCAAACCCAAAAAAAGAGATCTTTAATACGACTCACTATAGGGCGAGCGCCGCCATGGAGTACCCATACGACGTACCAGATTACGCTCATATGGCCATGGAGGCCAGTGAATTCCACCCGGGTGGGCATCGATACGGGATCCATCGAGCTCGAGCTGCAGATGAATCGTAGATACTGAAAAACCCCGCAAGTTCACTTCAACTGTGCATCGTGCACCATCTCAATTTCTTTCATTTATACATCGTTTTGCCTTCTTTTATGTAACTATACTCCTCTAAGTTTCAATCTTGGCCATGTAACCTCTGATCTATAGAATTTTTTAAATGACTAGAATTAATGCCCATCTTTTTTTTGGACCTAAATTCTTCATGAAAATATATTACGAGGGCTTATTCAGAAGCTTTGGACTTCTTCGCCAGAGGTTTGGTCAAGTCTCCAATCAAGGTTGTCGGCTTGTCTACCTTGCCAGAAATTTACGAAAAGATGGAAAAGGGTCAAATCGTTGGTAGATACGTTGTTGACACTTCTAAATAAGCGAATTTCTTATGATTTATGATTTTTATTATTAAATAAGTTATAAAAAAAATAAGTGTATACAAATTTTAAAGTGACTCTTAGGTTTTAAAACGAAAATTCTTATTCTTGAGTAACTCTTTCCTGTAGGTCAGGTTGCTTTCTCAGGTATAGCATGAGGTCGCTCTTATTGACCACACCTCTACCGGCCGGTCGAAATTCCCCTACCCTATGAACATATTCCATTTTGTAATTTCGTGTCGTTTCTATTATGAATTTCATTTATAAAGTTTATGTACAAATATCATAAAAAAAGAGAATCTTTTTAAGCAAGGATTTTCTTAACTTCTTCGGCGACAGCATCACCGACTTCGGTGGTACTGTTGGAACCACCTAAATCACCAGTTCTGATACCTGCATCCAAAACCTTTTTAACTGCATCTTCAATGGCCTTACCTTCTTCAGGCAAGTTCAATGACAATTTCAACATCATTGCAGCAGACAAGATAGTGGCGATAGGGTTGACCTTATTCTTTGGCAAATCTGGAGCAGAACCGTGGCATGGTTCGTACAAACCAAATGCGGTGTTCTTGTCTGGCAAAGAGGCCAAGGACGCAGATGGCAACAAACCCAAGGAACCTGGGATAACGGAGGCTTCATCGGAGATGATATCACCAAACATGTTGCTGGTGATTATAATACCATTTAGGTGGGTTGGGTTCTTAACTAGGATCATGGCGGCAGAATCAATCAATTGATGTTGAACCTTCAATGTAGGAAATTCGTTCTTGATGGTTTCCTCCACAGTTTTTCTCCATAATCTTGAAGAGGCCAAAACATTAGCTTTATCCAAGGACCAAATAGGCAATGGTGGCTCATGTTGTAGGGCCATGAAAGCGGCCATTCTTGTGATTCTTTGCACTTCTGGAACGGTGTATTGTTCACTATCCCAAGCGACACCATCACCATCGTCTTCCTTTCTCTTACCAAAGTAAATACCTCCCACTAATTCTCTGACAACAACGAAGTCAGTACCTTTAGCAAATTGTGGCTTGATTGGAGATAAGTCTAAAAGAGAGTCGGATGCAAAGTTACATGGTCTTAAGTTGGCGTACAATTGAAGTTCTTTACGGATTTTTAGTAAACCTTGTTCAGGTCTAACACTACCTGTACCCCATTTAGGACCACCCACAGCACCTAACAAAACGGCATCAGCCTTCTTGGAGGCTTCCAGCGCCTCATCTGGAAGTGGGACACCTGTAGCTTCGATAGCAGCACCACCAATTAAATGATTTTCGAAATCGAACTTGACATTGGAACGAACATCAGAAATAGCTTTAAGAACCTTAATGGCTTCGGCTGTGATTTCTTGACCAACGTGGTCACCTGGCAAAACGACGATCTTCTTAGGGGCAGACATTAGAATGGTATATCCTTGAAATATATATATATATTGCTGAAATGTAAAAGGTAAGAAAAGTTAGAAAGTAAGACGATTGCTAACCACCTATTGGAAAAAACAATAGGTCCTTAAATAATATTGTCAACTTCAAGTATTGTGATGCAAGCATTTAGTCATGAACGCTTCTCTATTCTATATGAAAAGCCGGTTCCGGCGCTCTCACCTTTCCTTTTTCTCCCAATTTTTCAGTTGAAAAAGGTATATGCGTCAGGCGACCTCTGAAATTAACAAAAAATTTCCAGTCATCGAATTTGATTCTGTGCGATAGCGCCCCTGTGTGTTCTCGTTATGTTGAGGAAAAAAATAATGGTTGCTAAGAGATTCGAACTCTTGCATCTTACGATACCTGAGTATTCCCACAGTTGGGGGATCTCGACTCTAGCTAGAGGATCAATTCGTAATCATGTCATAGCTGTTTCCTGTGTGAAATTGTTATCCGCTCACAATTCCACACAACATACGAGCCGGAAGCATAAAGTGTAAAGCCTGGGGTGCCTAATGAGTGAGCTAACTCACATTAATTGCGTTGCGCTCACTGCCCGCTTTCCAGTCGGGAAACCTGTCGTGCCAGCTGATAACTTCGTATAATGTATGCTATACGAAGTTATTAGGTCTGAAGAGGAGTTTACGTCCAGCCAAGCTAGCTTGGCTGCAGGTCGAGCGGCCGCGATCCGGAACCCTTAATATAACTTCGTATAATGTATGCTATACGAAGTTATCAGCTGCATTAATGAATCGGCCAACGCGCGGGGAGAGGCGGTTTGCGTATTGGGCGCTCTTCCGCTTCCTCGCTCACTGACTCGCTGCGCTCGGTCGTTCGGCTGCGGCGAGCGGTATCAGCTCACTCAAAGGCGGTAATACGGTTATCCACAGAATCAGGGGATAACGCAGGAAAGAACATGTGAGCAAAAGGCCAGCAAAAGGCCAGGAACCGTAAAAAGGCCGCGTTGCTGGCGTTTTTCCATAGGCTCCGCCCCCCTGACGAGCATCACAAAAATCGACGCTCAAGTCAGAGGTGGCGAAACCCGACAGGACTATAAAGATACCAGGCGTTTCCCCCTGGAAGCTCCCTCGTGCGCTCTCCTGTTCCGACCCTGCCGCTTACCGGATACCTGTCCGCCTTTCTCCCTTCGGGAAGCGTGGCGCTTTCTCATAGCTCACGCTGTAGGTATCTCAGTTCGGTGTAGGTCGTTCGCTCCAAGCTGGGCTGTGTGCACGAACCCCCCGTTCAGCCCGACCGCTGCGCCTTATCCGGTAACTATCGTCTTGAGTCCAACCCGGTAAGACACGACTTATCGCCACTGGCAGCAGCCACTGGTAACAGGATTAGCAGAGCGAGGTATGTAGGCGGTGCTACAGAGTTCTTGAAGTGGTGGCCTAACTACGGCTACACTAGAAGAACAGTATTTGGTATCTGCGCTCTGCTGAAGCCAGTTACCTTCGGAAAAAGAGTTGGTAGCTCTTGATCCGGCAAACAAACCACCGCTGGTAGCGGTGGTTTTTTTGTTTGCAAGCAGCAGATTACGCGCAGAAAAAAAGGATCTCAAGAAGATCCTTTGATCTTTTCTACGGGGTCTGACGCTCAGTGGAACGAAAACTCACGTTAAGGGATTTTGGTCATGAGATTATCAAAAAGGATCTTCACCTAGATCCTTTTAAATTAAAAATGAAGTTTTAAATCAATCTAAAGTATATATGAGTAAACTTGGTCTGACAGTTACCAATGCTTAATCAGTGAGGCACCTATCTCAGCGATCTGTCTATTTCGTTCATCCATAGTTGCCTGACTCCCCGTCGTGTAGATAACTACGATACGGGAGGGCTTACCATCTGGCCCCAGTGCTGCAATGATACCGCGAGACCCACGCTCACCGGCTCCAGATTTATCAGCAATAAACCAGCCAGCCGGAAGGGCCGAGCGCAGAAGTGGTCCTGCAACTTTATCCGCCTCCATCCAGTCTATTAATTGTTGCCGGGAAGCTAGAGTAAGTAGTTCGCCAGTTAATAGTTTGCGCAACGTTGTTGCCATTGCTACAGGCATCGTGGTGTCACGCTCGTCGTTTGGTATGGCTTCATTCAGCTCCGGTTCCCAACGATCAAGGCGAGTTACATGATCCCCCATGTTGTGCAAAAAAGCGGTTAGCTCCTTCGGTCCTCCGATCGTTGTCAGAAGTAAGTTGGCCGCAGTGTTATCACTCATGGTTATGGCAGCACTGCATAATTCTCTTACTGTCATGCCATCCGTAAGATGCTTTTCTGTGACTGGTGAGTACTCAACCAAGTCATTCTGAGAATAGTGTATGCGGCGACCGAGTTGCTCTTGCCCGGCGTCAATACGGGATAATACCGCGCCACATAGCAGAACTTTAAAAGTGCTCATCATTGGAAAACGTTCTTCGGGGCGAAAACTCTCAAGGATCTTACCGCTGTTGAGATCCAGTTCGATGTAACCCACTCGTGCACCCAACTGATCTTCAGCATCTTTTACTTTCACCAGCGTTTCTGGGTGAGCAAAAACAGGAAGGCAAAATGCCGCAAAAAAGGGAATAAGGGCGACACGGAAATGTTGAATACTCATACTCTTCCTTTTTCAATATTATTGAAGCATTTATCAGGGTTATTGTCTCATGAGCGGATACATATTTGAATGTATTTAGAAAAATAAACAAATAGGGGTTCCGCGCACATTTCCCCGAAAAGTGCCACCTGACGTCTAAGAAACCATTATTATCATGACATTAACCTATAAAAATAGGCGTATCACGAGGCCCTTTCGTCTCGCGCGTTTCGGTGATGACGGTGAAAACCTCTGACACATGCAGCTCCCGGAGACGGTCACAGCTTGTCTGTAAGCGGATGCCGGGAGCAGACAAGCCCGTCAGGGCGCGTCAGCGGGTGTTGGCGGGTGTCGGGGCTGGCTTAACTATGCGGCATCAGAGCAGATTGTACTGAGAGTGCACCATAACGCATTTAAGCATAAACACGCACTATGCCGTTCTTCTCATGTATATATATATACAGGCAACACGCAGATATAGGTGCGACGTGAACAGTGAGCTGTATGTGCGCAGCTCGCGTTGCATTTTCGGAAGCGCTCGTTTTCGGAAACGCTTTGAAGTTCCTATTCCGAAGTTCCTATTCTCTAGCTAGAAAGTATAGGAACTTCAGAGCGCTTTTGAAAACCAAAAGCGCTCTGAAGACGCACTTTCAAAAAACCAAAAACGCACCGGACTGTAACGAGCTACTAAAATATTGCGAATACCGCTTCCACAAACATTGCTCAAAAGTATCTCTTTGCTATATATCTCTGTGCTATATCCCTATATAACCTACCCATCCACCTTTCGCTCCTTGAACTTGCATCTAAACTCGACCTCTACATTTTTTATGTTTATCTCTAGTATTACTCTTTAGACAAAAAAATTGTAGTAAGAACTATTCATAGAGTGAATCGAAAACAATACGAAAATGTAAACATTTCCTATACGTAGTATATAGAGACAAAATAGAAGAAACCGTTCATAATTTTCTGACCAATGAAGAATCATCAACGCTATCACTTTCTGTTCACAAAGTATGCGCAATCCACATCGGTATAGAATATAATCGGGGATGCCTTTATCTTGAAAAAATGCACCCGCAGCTTCGCTAGTAATCAGTAAACGCGGGAAGTGGAGTCAGGCTTTTTTTATGGAAGAGAAAATAGACACCAAAGTAGCCTTCTTCTAACCTTAACGGACCTACAGTGCAAAAAGTTATCAAGAGACTGCATTATAGAGCGCACAAAGGAGAAAAAAAGTAATCTAAGATGCTTTGTTAGAAAAATAGCGCTCTCGGGATGCATTTTTGTAGAACAAAAAAGAAGTATAGATTCTTTGTTGGTAAAATAGCGCTCTCGCGTTGCATTTCTGTTCTGTAAAAATGCAGCTCAGATTCTTTGTTTGAAAAATTAGCGCTCTCGCGTTGCATTTTTGTTTTACAAAAATGAAGCACAGATTCTTCGTTGGTAAAATAGCGCTTTCGCGTTGCATTTCTGTTCTGTAAAAATGCAGCTCAGATTCTTTGTTTGAAAAATTAGCGCTCTCGCGTTGCATTTTTGTTCTACAAAATGAAGCACAGATGCTTCGTTGCT
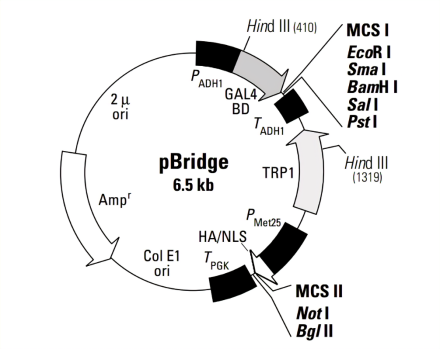
2.Vector Name: pBridge
Promoter: ADH1
Origin of Replication: 2μori, ColE1 ori
Vector Size: 6526bp
5' Sequencing Primer and Sequence: 5'-TCATCGGAAGAGAGTAGT-3'
3' Sequencing Primer and Sequence: 5'-AATTTATTTCTTTTCGGATAA-3'
Vector Resistance: Ampicillin
Selection Marker: TRP1
>pBridge vector sequence
GCTTGCATGCAACTTCTTTTCTTTTTTTTTCTTTTCTCTCTCCCCCGTTGTTGTCTCACCATATCCGCAATGACAAAAAAATGATGGAAGACACTAAAGGAAAAAATTAACGACAAAGACAGCACCAACAGATGTCGTTGTTCCAGAGCTGATGAGGGGTATCTCGAAGCACACGAAACTTTTTCCTTCCTTCATTCACGCACACTACTCTCTAATGAGCAACGGTATACGGCCTTCCTTCCAGTTACTTGAATTTGAAATAAAAAAAAGTTTGCTGTCTTGCTATCAAGTATAAATAGACCTGCAATTATTAATCTTTTGTTTCCTCGTCATTGTTCTCGTTCCCTTTCTTCCTTGTTTCTTTTTCTGCACAATATTTCAAGCTATACCAAGCATACAATCAACTCCAAGCTTGAAGCAAGCCTCCTGAAAGATGAAGCTACTGTCTTCTATCGAACAAGCATGCGATATTTGCCGACTTAAAAAGCTCAAGTGCTCCAAAGAAAAACCGAAGTGCGCCAAGTGTCTGAAGAACAACTGGGAGTGTCGCTACTCTCCCAAAACCAAAAGGTCTCCGCTGACTAGGGCACATCTGACAGAAGTGGAATCAAGGCTAGAAAGACTGGAACAGCTATTTCTACTGATTTTTCCTCGAGAAGACCTTGACATGATTTTGAAAATGGATTCTTTACAGGATATAAAAGCATTGTTAACAGGATTATTTGTACAAGATAATGTGAATAAAGATGCCGTCACAGATAGATTGGCTTCAGTGGAGACTGATATGCCTCTAACATTGAGACAGCATAGAATAAGTGCGACATCATCATCGGAAGAGAGTAGTAACAAAGGTCAAAGACAGTTGACTGTATCGCCGGAATTCCCGGGGATCCGTCGACCTGCAGCCAAGCTAATTCCGGGCGAATTTCTTATGATTTATGATTTTTATTATTAAATAAGTTATAAAAAAAATAAGTGTATACAAATTTTAAAGTGACTCTTAGGTTTTAAAACGAAAATTCTTATTCTTGAGTAACTCTTTCCTGTAGGTCAGGTTGCTTTCTCAGGTATAGCATGAGGTCGCTCTTATTGACCACACCTCTACCGGCATGCCGGCAAGTGCACAAACAATACTTAAATAAATACTACTCAGTAATAACCTATTTCTTAGCATTTTTGACGAAATTTGCTATTTTGTTAGAGTCTTTTACACCATTTGTCTCCACACCTCCGCTTACATCAACACCAATAACGCCATTTAATCTAAGCGCATCACCAACATTTTCTGGCGTCAGTCCACCAGCTAACATAAAATGTAAGCTTTCGGGGCTCTCTTGCCTTCCAACCCAGTCAGAAATCGAGTTCCAATCCAAAAGTTCACCTGTCCCACCTGCTTCTGAATCAAACAAGGGAATAAACGAATGAGGTTTCTGTGAAGCTGCACTGAGTAGTATGTTGCAGTCTTTTGGAAATACGAGTCTTTTAATAACTGGCAAACCGAGGAACTCTTGGTATTCTTGCCACGACTCATCTCCATGCAGTTGGACGATATCAATGCCGTAATCATTGACCAGAGCCAAAACATCCTCCTTAGGTTGATTACGAAACACGCCAACCAAGTATTTCGGAGTGCCTGAACTATTTTTATATGCTTTTACAAGACTTGAAATTTTCCTTGCAATAACCGGGTCAATTGTTCTCTTTCTATTGGGCACACATATAATACCCAGCAAGTCAGCATCGGAATCTAGAGCACATTCTGCGGCCTCTGTGCTCTGCAAGCCGCAAACTTTCACCAATGGACCAGAACTACCTGTGAAATTAATAACAGACATACTCCAAGCTGCCTTTGTGTGCTTAATCACGTATACTCACGTGCTCAATAGTCACCAATGCCCTCCCTCTTGGCCCTCTCCTTTTCTTTTTTCGACCGAATTAATTCGTAATCATGTCATAGCTGTTTCCTGTGTGAAATTGTTATCCGCTCACAATTCCACACAACATACGAGCCGGAAGCATAAAGTGTAAAGCCTGGGGTGCCTAATGAGTGAGCTAACTCACATTAATTGCGTTGCGCTCACTGCCCGCTTTCCAGTCGGGAAACCTGTCGTGCCAGGAAGATCCGAGGCCTAGCTTCTAATTCTTCCAACATACAATGGGAGTTTGGCCGAGTGGTTTAAGGCGTCAGATTTAGGTGGATTTAACCTCTAAAATCTCTGATATCTTCGGATGCAAGGGTTCGAATCCCTTAGCTCTCATTATTTTTTGCTTTTTCTCTTGAGGTCACATGATCGCAAAATGGCAAATGGCACGTGAAGCTGTCGATATTGGGGAACTGTGGTGGTTGGCAAATGACTAATTAAGTTAGTCAAGGCGCCATCCTCATGAAAACTGTGTAACATAATAACCGAAGTGTCGAAAAGGTGGCACCTTGTCCAATTGAACACGCTCGATGAAAAAAATAAGATATATATAAGGTTAAGTAAAGCGTCTGTTAGAAAGGAAGTTTTTCCTTTTTCTTGCTCTCTTGTCTTTTCATCTACTATTTCCTTCGTGTAATACAGGGTCGTCAGATACATAGATACAATTCTATTACCCCCATCCATACAATGGGCCATATGGCTTCTAGCTATCCTTATGACGTGCCTGACTATGCCAGCCTGGGAGGACCTTCTAGTCCTAAGAAGAAGAGAAAGGTGGCGGCCGCATTAGCCCGAAGATCTTCGGGCTGATCTCCCATGTCTCTACTGGTGGTGGTGCTTCTTTGGAATTATTGGAAGGTAAGGAATTGCCAGGTGTTGCTTTCTTATCCGAAAAGAAATAAATTGAATTGAATTGAAATCGATAGATCAATTTTTTTCTTTTCTCTTTCCCCATCCTTTACGCTAAAATAATAGTTTATTTTATTTTTTGAATATTTTTTATTTATATACGTATATATAGACTATTATTTATCTTTTAATGATTATTAAGATTTTTATTAAAAAAAAATTCGCTCCTCTTTTAATGCCTTTATGCAGTTTTTTTTTCCCATTCGATATTTCTATGTTCGGGTTCAGCGTATTTTAAGTTTAATAACTCGAAAATTCTGCGTTCGTTAAAGCTAGGCCTCGGATCTTCCTGCATTAATGAATCGGCCAACGCGCGGGGAGAGGCGGTTTGCGTATTGGGCGCTCTTCCGCTTCCTCGCTCACTGACTCGCTGCGCTCGGTCGTTCGGCTGCGGCGAGCGGTATCAGCTCACTCAAAGGCGGTAATACGGTTATCCACAGAATCAGGGGATAACGCAGGAAAGAACATGTGAGCAAAAGGCCAGCAAAAGGCCAGGAACCGTAAAAAGGCCGCGTTGCTGGCGTTTTTCCATAGGCTCCGCCCCCCTGACGAGCATCACAAAAATCGACGCTCAAGTCAGAGGTGGCGAAACCCGACAGGACTATAAAGATACCAGGCGTTTCCCCCTGGAAGCTCCCTCGTGCGCTCTCCTGTTCCGACCCTGCCGCTTACCGGATACCTGTCCGCCTTTCTCCCTTCGGGAAGCGTGGCGCTTTCTCATAGCTCACGCTGTAGGTATCTCAGTTCGGTGTAGGTCGTTCGCTCCAAGCTGGGCTGTGTGCACGAACCCCCCGTTCAGCCCGACCGCTGCGCCTTATCCGGTAACTATCGTCTTGAGTCCAACCCGGTAAGACACGACTTATCGCCACTGGCAGCAGCCACTGGTAACAGGATTAGCAGAGCGAGGTATGTAGGCGGTGCTACAGAGTTCTTGAAGTGGTGGCCTAACTACGGCTACACTAGAAGAACAGTATTTGGTATCTGCGCTCTGCTGAAGCCAGTTACCTTCGGAAAAAGAGTTGGTAGCTCTTGATCCGGCAAACAAACCACCGCTGGTAGCGGTGGTTTTTTTGTTTGCAAGCAGCAGATTACGCGCAGAAAAAAAGGATCTCAAGAAGATCCTTTGATCTTTTCTACGGGGTCTGACGCTCAGTGGAACGAAAACTCACGTTAAGGGATTTTGGTCATGAGATTATCAAAAAGGATCTTCACCTAGATCCTTTTAAATTAAAAATGAAGTTTTAAATCAATCTAAAGTATATATGAGTAAACTTGGTCTGACAGTTACCAATGCTTAATCAGTGAGGCACCTATCTCAGCGATCTGTCTATTTCGTTCATCCATAGTTGCCTGACTCCCCGTCGTGTAGATAACTACGATACGGGAGGGCTTACCATCTGGCCCCAGTGCTGCAATGATACCGCGAGACCCACGCTCACCGGCTCCAGATTTATCAGCAATAAACCAGCCAGCCGGAAGGGCCGAGCGCAGAAGTGGTCCTGCAACTTTATCCGCCTCCATCCAGTCTATTAATTGTTGCCGGGAAGCTAGAGTAAGTAGTTCGCCAGTTAATAGTTTGCGCAACGTTGTTGCCATTGCTACAGGCATCGTGGTGTCACGCTCGTCGTTTGGTATGGCTTCATTCAGCTCCGGTTCCCAACGATCAAGGCGAGTTACATGATCCCCCATGTTGTGCAAAAAAGCGGTTAGCTCCTTCGGTCCTCCGATCGTTGTCAGAAGTAAGTTGGCCGCAGTGTTATCACTCATGGTTATGGCAGCACTGCATAATTCTCTTACTGTCATGCCATCCGTAAGATGCTTTTCTGTGACTGGTGAGTACTCAACCAAGTCATTCTGAGAATAGTGTATGCGGCGACCGAGTTGCTCTTGCCCGGCGTCAATACGGGATAATACCGCGCCACATAGCAGAACTTTAAAAGTGCTCATCATTGGAAAACGTTCTTCGGGGCGAAAACTCTCAAGGATCTTACCGCTGTTGAGATCCAGTTCGATGTAACCCACTCGTGCACCCAACTGATCTTCAGCATCTTTTACTTTCACCAGCGTTTCTGGGTGAGCAAAAACAGGAAGGCAAAATGCCGCAAAAAAGGGAATAAGGGCGACACGGAAATGTTGAATACTCATACTCTTCCTTTTTCAATATTATTGAAGCATTTATCAGGGTTATTGTCTCATGAGCGGATACATATTTGAATGTATTTAGAAAAATAAACAAATAGGGGTTCCGCGCACATTTCCCCGAAAAGTGCCACCTGACGTCTAAGAAACCATTATTATCATGACATTAACCTATAAAAATAGGCGTATCACGAGGCCCTTTCGTCTCGCGCGTTTCGGTGATGACGGTGAAAACCTCTGACACATGCAGCTCCCGGAGACGGTCACAGCTTGTCTGTAAGCGGATGCCGGGAGCAGACAAGCCCGTCAGGGCGCGTCAGCGGGTGTTGGCGGGTGTCGGGGCTGGCTTAACTATGCGGCATCAGAGCAGATTGTACTGAGAGTGCACCATAACGCATTTAAGCATAAACACGCACTATGCCGTTCTTCTCATGTATATATATATACAGGCAACACGCAGATATAGGTGCGACGTGAACAGTGAGCTGTATGTGCGCAGCTCGCGTTGCATTTTCGGAAGCGCTCGTTTTCGGAAACGCTTTGAAGTTCCTATTCCGAAGTTCCTATTCTCTAGCTAGAAAGTATAGGAACTTCAGAGCGCTTTTGAAAACCAAAAGCGCTCTGAAGACGCACTTTCAAAAAACCAAAAACGCACCGGACTGTAACGAGCTACTAAAATATTGCGAATACCGCTTCCACAAACATTGCTCAAAAGTATCTCTTTGCTATATATCTCTGTGCTATATCCCTATATAACCTACCCATCCACCTTTCGCTCCTTGAACTTGCATCTAAACTCGACCTCTACATTTTTTATGTTTATCTCTAGTATTACTCTTTAGACAAAAAAATTGTAGTAAGAACTATTCATAGAGTGAATCGAAAACAATACGAAAATGTAAACATTTCCTATACGTAGTATATAGAGACAAAATAGAAGAAACCGTTCATAATTTTCTGACCAATGAAGAATCATCAACGCTATCACTTTCTGTTCACAAAGTATGCGCAATCCACATCGGTATAGAATATAATCGGGGATGCCTTTATCTTGAAAAAATGCACCCGCAGCTTCGCTAGTAATCAGTAAACGCGGGAAGTGGAGTCAGGCTTTTTTTATGGAAGAGAAAATAGACACCAAAGTAGCCTTCTTCTAACCTTAACGGACCTACAGTGCAAAAAGTTATCAAGAGACTGCATTATAGAGCGCACAAAGGAGAAAAAAAGTAATCTAAGATGCTTTGTTAGAAAAATAGCGCTCTCGGGATGCATTTTTGTAGAACAAAAAAGAAGTATAGATTCTTTGTTGGTAAAATAGCGCTCTCGCGTTGCATTTCTGTTCTGTAAAAATGCAGCTCAGATTCTTTGTTTGAAAAATTAGCGCTCTCGCGTTGCATTTTTGTTTTACAAAAATGAAGCACAGATTCTTCGTTGGTAAAATAGCGCTTTCGCGTTGCATTTCTGTTCTGTAAAAATGCAGCTCAGATTCTTTGTTTGAAAAATTAGCGCTCTCGCGTTGCATTTTTGTTCTACAAAATGAAGCACAGATGCTTCGTT
- Wang, S(2024). Yeast Three-Hybrid Screening and Validation Experiment Protocol. Bio-protocol Preprint. bio-protocol.org/prep2735.
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link
