Advanced Search
Nuclei Isolation from Murine and Human Periosteum for Transcriptomic Analyses
Last updated date: Oct 18, 2024 Views: 208 Forks: 0
NUCLEI ISOLATION FROM MURINE AND HUMAN PERIOSTEUM FOR TRANSCRIPTOMIC ANALYSES
Simon Perrin1, Cassandre Goachet1, Maria Ethel1, Yasmine Hachemi1 and Céline Colnot 1, *
1Univ Paris Est Creteil, INSERM, IMRB, F-94010 Creteil, France
*For correspondence: celine.colnot@inserm.fr
[Abstract]
Bone repair is a complex regenerative process relying on skeletal stem/progenitor cells (SSPCs) recruited predominantly from the periosteum. Activation and differentiation of periosteal SSPCs occurs in an heterogeneous environment, rising the need of using single cell/nucleus transcriptomics in order to decipher the response of the periosteum to injury. Yet, enzymatic cell dissociation can induce a stress response affecting the transcriptome and lead to overrepresentation of certain cell types (i.e. immune and endothelial cells) and low coverage of other cell types of interest. To counteract these limitations, we optimized a protocol to isolate nuclei directly from the intact periosteum and from the fracture callus to perform single-nucleus RNA sequencing (snRNAseq). This protocol is adapted for fresh murine periosteum, fracture callus and for frozen human periosteum. Nuclei are isolated using mechanical extraction combined with fluorescence-based nuclei sorting to obtain high-quality nucleus suspension. This protocol allows to capture the full diversity of cell types in the periosteum and the fracture environment to better reflect the in vivo tissue composition.
Graphical abstract
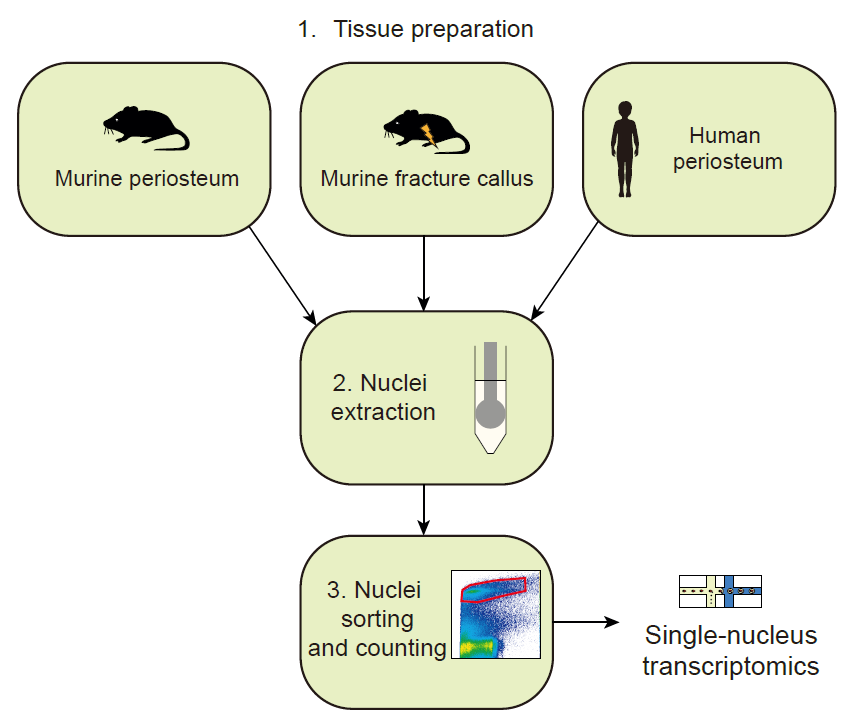
Key Features
Allows the isolation of nuclei with high quality RNA for transcriptomic analyses.
Can be adapted to be used on fresh and frozen tissue.
Optimized for human and murine periosteum.
Keywords: Nuclei isolation, Single-nucleus RNA-seq, Periosteum, Fracture callus, Fluorescence-activated nuclei sorting
[Background]
The periosteum is a thin and heterogenous tissue covering the outer surface of bone and contains skeletal stem/progenitor cells (SSPCs) essential for bone repair. Following a bone fracture, periosteal SSPCs are activated and differentiate to form cartilage and bone cells (Duchamp de Lageneste et al., 2018; Jeffery et al., 2022; Perrin et al., 2023; Perrin & Colnot, 2022). SSPCs differentiate in a highly complex and dynamic environment marked by the presence of many cell types, including immune cells, endothelial cells and cells from the nervous system (Hachemi et al., 2024; Perrin et al., 2023). In order to decipher the heterogeneity of the periosteum and the response of these different cell types to bone fracture, we aimed to perform single-nucleus/cell transcriptomics from the periosteum and callus tissues. Cell isolation using enzymatic digestion is commonly used, yet there are several disadvantages in this method. First, the enzymatic tissue digestion can favor the representation of certain cell types. It usually leads to the overrepresentation of cell types loosely attached to the matrix, such as immune cells, and to the limited coverage of cell types embedded in the matrix, including osteoblasts, osteoclasts and Schwann cells. Also, FACS-sorting and single cell transcriptomic techniques have constrains in terms of cell size that can be processed. The fracture callus contains large cells such as hypertrophic chondrocytes and osteoclasts, that are not be compatible with these techniques. In addition, while cell digestion can be performed on freshly dissected tissue, it cannot be considered for frozen tissue. To counteract these limitations, we optimized and tested a protocol to isolate nuclei from fresh and frozen tissue to perform single-nucleus RNA sequencing (snRNAseq). SnRNAseq was shown to produce similar results as single-cell RNA sequencing (scRNAseq), but can capture a greater diversity of cell types compared to scRNAseq (Ding et al., 2020; Machado et al., 2021; Selewa et al., 2020; Wen et al., 2022; Wu et al., 2019). In addition, enzymatic digestion needed for scRNAseq induces a stress response, leading to transcriptomic changes that are overcome by snRNAseq (Machado et al., 2021). Here, we detail 3 distinct ways of using our protocol in order to isolate nuclei (i) from fresh murine periosteum, (ii) from fresh murine fracture hematoma/callus and (iii) from frozen human periosteum. While tissue preparation can change between the different types of samples, the nuclei extraction protocol is common and based on mechanical cell lysis. Nuclei extraction is followed by fluorescence-activated nuclei sorting to eliminate the yields of cellular and matrix debris in the suspension. Overall, this protocol is fast, easy-to-use and is currently the most adequate method to capture the periosteal heterogeneity in vivo.
Materials and Reagents
Biological Materials
8 to 12 week-old mice in the C57BL6 background
Periosteum from patients
Reagents
- S&T Microsurgical Needles with Suture Thread Attached (Fine Science Tools, catalog number: 12051-08)
DMEM medium (Life Technologies, catalog number : 11966025)
HEPES buffer (Thermofisher, catalog number: 15630056)
Penicilin/streptomycin (Life Technologies, catalog number : 15140122)
PBS RNAse-free (Thermofisher, catalog number: AM9624)
Ethanol Absolute (VWR, catalog number : 20821.365)
Nuclei lysis buffer (Sigma-Aldrich, catalog number: NUC101-1KT)
Bovine Serum Albumin (Merck, catalog number: A2153)
RNAse inhibitor (Roche, catalog number: 3335399001)
DNase/RNase-Free distilled water, UltraPure (Life Technologies, catalog number : 10977049)
SYTOX™ AADvanced™ Dead Cell Stain Kit (Thermofisher, catalog number: S10349)
DAPI (Life Technologies, catalog number : D3571)
Buprenorphine (Centravet, catalog number: BUP001)
Atipamezole (Centravet, catalog number: ANT201)
Ketamine (Centravet, catalog number: KET205)
Medetomidine (Centravet, catalog number: DOM003)
Solutions
Human sample collection medium
70% ethanol
Nuclei buffer
DAPI solution
Recipes
Human sample collection medium
Reagent | Final concentration | Amount |
DMEM | 1X | 44.5 ml |
HEPES | 10% | 5 ml |
Pen/strep | 1% | 0.5 ml |
Total | n/a | 50 ml |
70% ethanol
Reagent | Final concentration | Amount |
Ethanol (absolute) | 70% | 700 ml |
H2O | n/a | 300 ml |
Total | n/a | 1000 ml |
Nuclei buffer
Reagent | Final concentration | Amount |
RNAse-free PBS (10X) | 1X | 0.5 ml |
Bovine Serum Albumin RNAse inhibitor (40 U/μl) RNAse-free water | 2% 0.2 U/μl n/a | 0.1 g 25 μl 4,5 ml |
Total | n/a | 5 ml |
DAPI buffer
Reagent | Final concentration | Amount |
Nuclei buffer | 1X | 99 μl |
DAPI (diluted 1/10) | 1/1000 | 1 μl |
Total | n/a | 100 μl |
Laboratory Supplies
Conical tubes, 15-ml and 50-ml (Falcon, catalog numbers: 352097 [15 ml] and 352070 [50 ml] or equivalent)
Eppendorf tubes 1.5 ml and 0.2 ml
Cryotube ClearLine® 2 ml (Dutscher, catalog number : 390701)
10-ml and 25 ml pipettes (Dutscher, catalog numbers: 357551 [10 ml] and 357535 [25 ml] or equivalent)
1ml, 200μl, 20μl, and 10μl pipette tips
Falcon 5ml round-bottomed polystyrene tubes (Corning, catalog number: 352235)
Sterilin™ Quickstart Universal Containers, PS, 30 ml (VWR, catalog number: 128AR/IRR, or equivalent)
Sterile scalpels (Dutscher, catalog number: 132622)
40-μm and 100-μm cell strainer (Fisher Scientific, catalog number: 352340 and 352360)
25 G needles (Terumo, catalog number: AN*2516R1)
1-ml syringes (Terumo, catalog number: SS+01H1)
Greiner Bio-One Petri dishes (bacterial dish; Dutscher, catalog number: 633185)
Kova® slides (Fisher Scientific, catalog number: 22-270141)
Liquid nitrogen
Ice
Equipment
Centrifuge with temperature control for Falcon 50 mL tube
Centrifuge with temperature control for Eppendorf 1.5 mL tube
BD Influx Cell Sorter, or equivalent
Zeiss Imager D1 AX10 light microscope (Carl Zeiss Microscopy), or equivalent
-80°C freezer
Sterile hood
Microvolume pipettes
Surgical forceps (Dumont AA Forceps; FST, catalog number: 11210-20, or equivalent)
Surgical scissors (Fine Scissors-ToughCut® 11 mm; FST, catalog number: 14058-11, or equivalent)
Dissecting Chisel (Fine Science Tools, catalog number: 10095-12)
Drill (Dremel, catalog number: 8050-15)
Drill bits (0.4 mm)
Heating pad (Harvard Apparatus, catalog number: 55-7033)
Trimmer (Kerbl, catalog number: GT416)
15 mL Dounce homogenizer with pestled (Sigma-Aldrich, catalog number: D9938)
Liquid nitrogen container
Ice container
Procedure
Prepare Material and Reagents
Set the centrifuge to 4°C and allow it to cool before use.
Prepare all solutions needed for the protocol.
Note : Prepare all reagents in RNAse free conditions. All solutions should be prepared fresh and kept on ice.
Prepare and annotate all tubes needed for the procedure.
Option I : Nuclei isolation from fresh murine periosteum
Sacrifice the mice by cervical dislocation (or any other appropriate method).
Note: as the periosteum is very thin in adult mice, we recommend using the tibias of at least 5 mice.
Note: all procedures involving animals must be approved by ethical committees.
Rinse the limbs with 70% ethanol.
Incise the skin and remove it entirely from the lower limbs. Disconnect the tibias from the limbs by cutting at the knee and ankle level. Place the tibias in a 100-mm sterile petri dish with RNAse-free chilled PBS on ice.
Remove the soft tissues surrounding the tibias using forceps and scissors.
Note: Remove soft tissue gently using scissors, as pulling out muscle could detach the periosteum from the cortex.
Note: Dissection steps can be performed under a binocular microscope to reduce the risk of contamination by surrounding tissue.
Cut the epiphyses of the tibia using scissors. Flush the bone marrow using RNAse-free PBS.
Collect the periosteum by scraping it from the cortex using dissecting Chisel. Collect the tissue in a 1.5ml Eppendorf containing lysis buffer and place it on ice.
Put the tissues in a 100-mm sterile petri dish with a drop of lysis buffer. Chop it with clean scissors until obtaining very small pieces.
Proceed immediately to nuclei extraction (Part E of this protocol)
Critical : do not wait to perform nuclei extraction, any time lost will impact RNA quality.
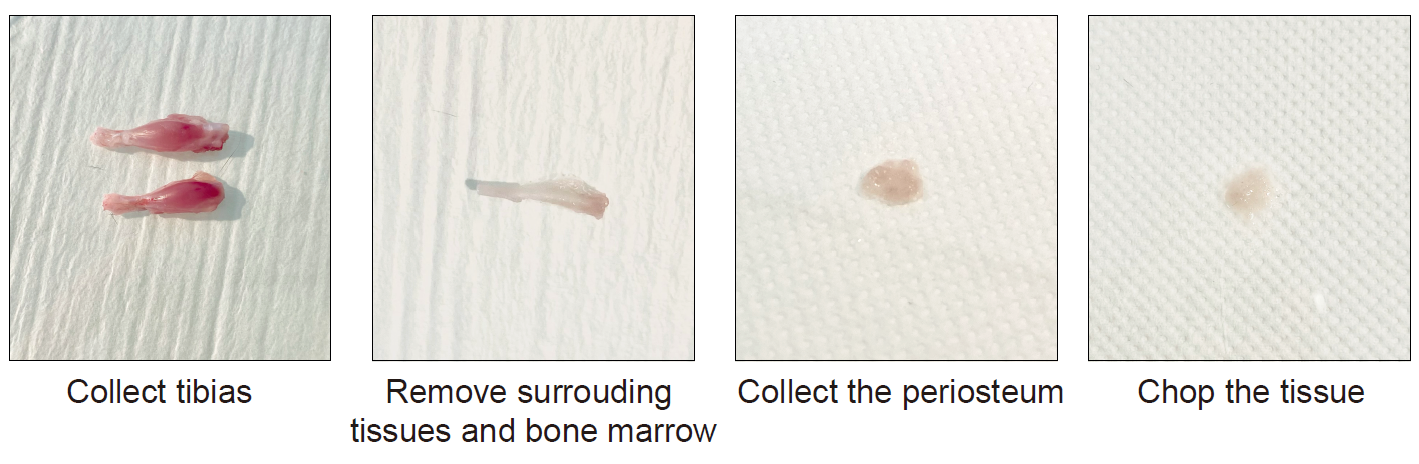
Figure 1 : Tissue processing of uninjured murine periosteum.
Option B : Isolation of fresh murine fracture hematoma/callus
Induce tibial fracture
Note: all procedures involving animals must be approved by ethical committees.
Note: we recommend using at least 5 mice for post-fracture day 1, and at least 3 mice for later time points to obtain sufficient number of nuclei and overcome interindividual variability.
Anesthetize the mice with intraperitoneal injection of 50 mg/kg ketamine and 1 mg/kg medetomidine. Inject 0.1 mg/kg of buprenorphine subcutaneously for analgesia.
After 15-30 min, if the quality of the anesthesia and analgesia is sufficient, shave the right limb and sanitize using skin disinfectant.
Note: The efficiency of anesthesia can be checked using foot pinching.
Perform a 2-cm incision on the skin along the tibia using a sterile scalpel and expose the tibial surface by gently separating the muscles from the bone surface.
Create 3 holes in the mid-diaphysis and aligned perpendicular to the tibial axis using a drill and a 0.4-mm drill bit.
Induce an osteotomy by cutting the bone along the 3 holes with scissors.
Close the skin wound using suture threads.
Revive the mice with an intraperitoneal injection of 1 mg/kg atipamezole and place it on a 37°C heating pad until revived.
Perform two additional subcutaneous injections of buprenorphine 0.1 mg/kg at 12 h and 24h post-surgery, and monitor the mice closely until callus tissue collection.
Collect tissue for nuclei extraction
Note: we tested our protocol on fracture tissue up to 7 days post-fracture. Later time points may need further optimization as the fracture callus becomes more ossified.
Sacrifice the mice by cervical dislocation (or any other appropriate method) and rinse the limbs with 70% ethanol.
Incise the skin and remove it entirely from the fractured limb. Disconnect the tibia from the limb by cutting at the knee and ankle level and place it in a 100-mm sterile petri dish with RNAse-free chilled PBS.
Note: proceed very gently to avoid separating the two segments of the tibia while dissecting as the fractured tibia is very fragile in the first days post-injury.
Remove the soft tissues surrounding the tibia using forceps and scissors.
Note: Remove soft tissue gently using scissors, as pulling out muscle could damage the fracture tissue.
Scrape the fracture hematoma/callus and the activated periosteum from the diaphysis using Dissecting Chisel. Collect the tissue in lysis buffer placed on ice.
Put the tissue with a drop of lysis buffer. Chop it with clean scissors until obtaining very small pieces.
Proceed immediately to nuclei extraction (Part E of this protocol)
Critical : do not wait to perform nuclei extraction, any time lost will impact RNA quality.
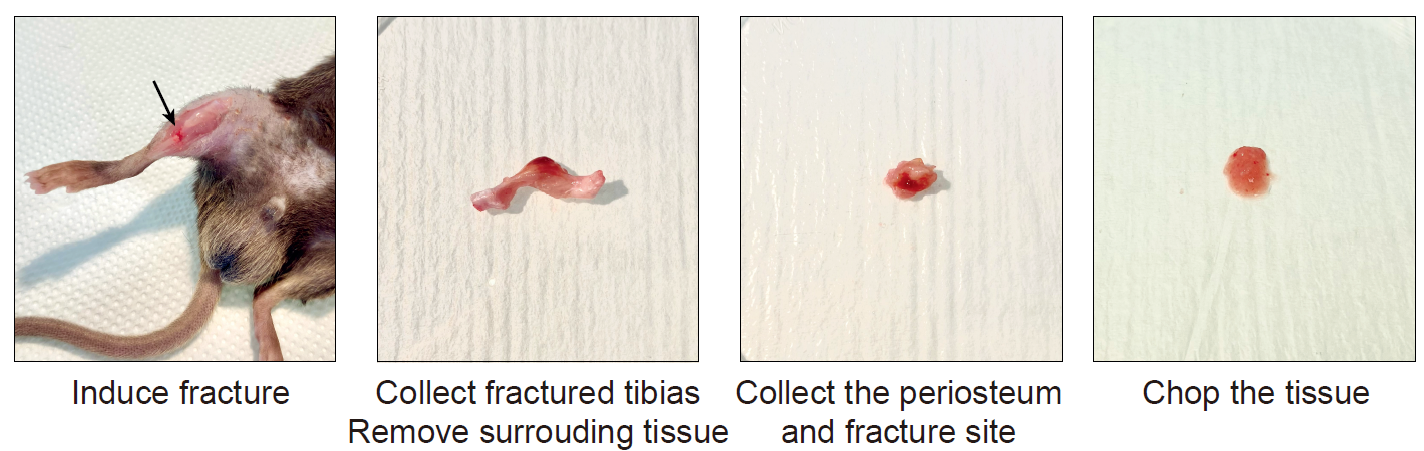
Figure 2 : Tissue processing of murine fracture callus and activated periosteum.
Option C : Isolation of frozen human periosteum
Collect and freeze the human periosteum
Place the sample in a tube containing at least 10 mL of human sample collection medium immediately after collection. Place at 4°C until processing.
Note: samples should be processed as early as possible after collection. In our experience, samples can be processed up to 3 hours after collection and be used for nuclei extraction.
Under a cell culture hood, remove the tissue from the tube and place it in a sterile petri dish. Wash the tissue with RNAse-free PBS to remove remaining medium and blood.
Using scissors, scalpel and tweezers, remove any muscle, fat tissue or damaged tissue.
Cut the periosteum in pieces of at least 0,5 x 0,5 cm. Place the pieces briefly in a dry petri dish to remove the medium from the tissue.
Note: we recommend storing larger pieces of tissue to obtain a higher number of nuclei after nuclei extraction. Store at least 3 pieces of tissues if possible.
Place each piece of periosteum in a freezing tube properly labeled.
Place the tubes directly in liquid nitrogen. Then, place the tubes at -80°C.
Caution : handle liquid nitrogen with care and follow safety rules.
Stop point : Samples can be stored at -80°C for a long time before processing. We have used samples up to 3 years after freezing.
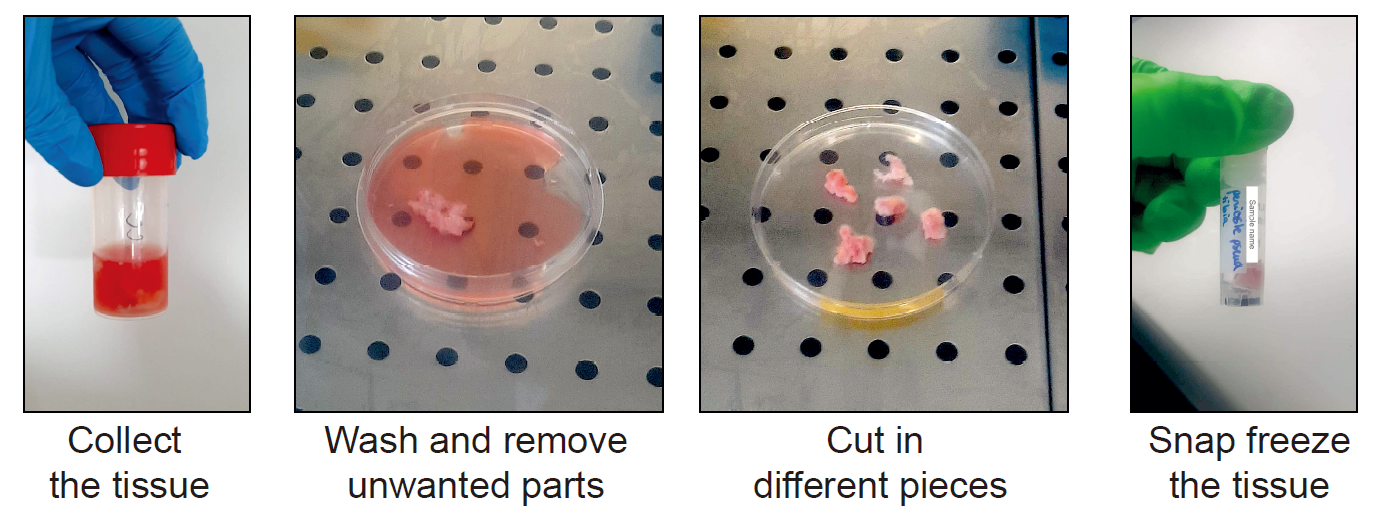
Figure 3 : Collection and freezing of the human periosteum
Check the RNA integrity
If possible, use one sample from the same batch to extract RNA and check that RNA integrity is above 6.5.
Note: we avoid using sample with RIN lower than 6,5 as it may lead to poor RNA quality after nuclei extraction
Prepare tissue for nuclei extraction
Take the sample from the -80°C freezer and put it on dry ice.
Critical point: Keep the tube on dry ice until using it for extraction. Do not allow the sample to thaw before placing it into the lysis buffer.
Put the frozen tissue on a RNAse-free petri dish. Add few drops of lysis buffer.
Chop the tissue with clean scissors until obtaining very small pieces.
Proceed immediately to nuclei extraction (Part E)
Critical : do not wait to perform nuclei extraction, any time lost will lead to a decrease in RNA quality.
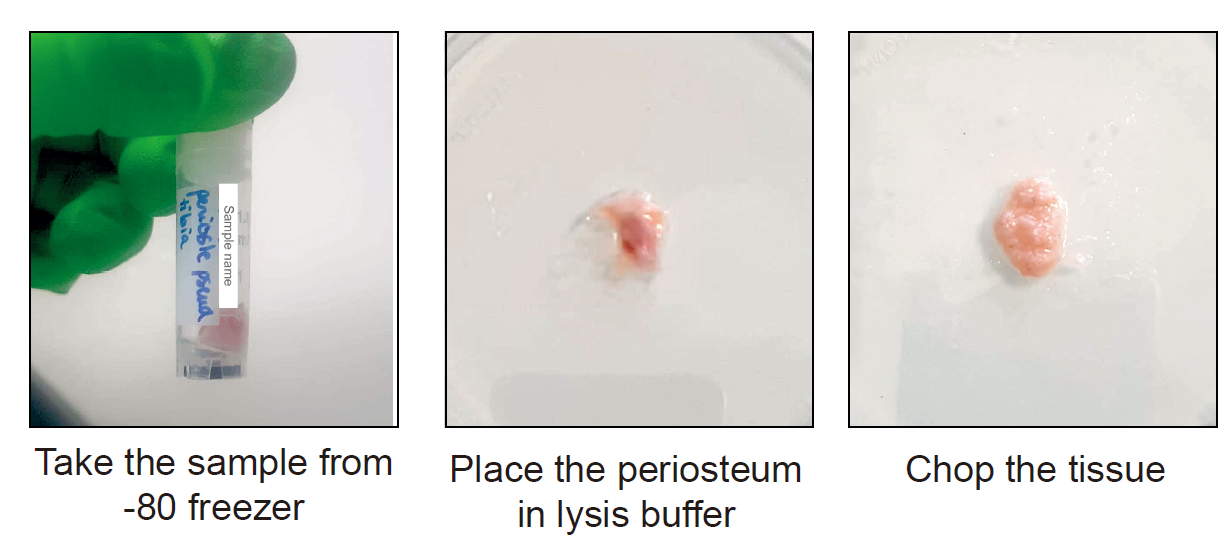
Figure 4 : Tissue processing of human periosteum
Nuclei extraction
Put the tissues prepared in part B, C or D in a glass douncer. Add up to 7 ml of lysis buffer.
Put the douncer on ice for 2 minutes.
While keeping the douncer on ice, lower and raise the pestle A gently 15-20 times, avoiding bubble formation.
Note: There shouldn’t be too much resistance while using pestles. Resistance can be due to presence of big pieces of tissue, that would require a better chopping before putting the tissue in the douncer, or to a high ratio tissue / lysis buffer, that would require putting less tissue or more lysis buffer if possible.
Lower and raise the pestle B gently up to 10 times.
Filter the suspension with a 100 μm cell strainer in a 50 mL conical tube. Collect the flow through and filter using a 40 μmcell strainer in a clean 50 mL tube.
Centrifuge at 500 × g for 5 min at 4°C. Carefully remove the supernatant without disturbing the pellet.
Add 1 ml of nuclei buffer. Resuspend the pellet and transfer in a RNAse-free Eppendorf tube.
Centrifuge at 500 × g for 5 min at 4°C. Carefully remove the supernatant without disturbing the pellet.
Resuspend in 200 μl of nuclei buffer. Proceed to nuclei sorting.
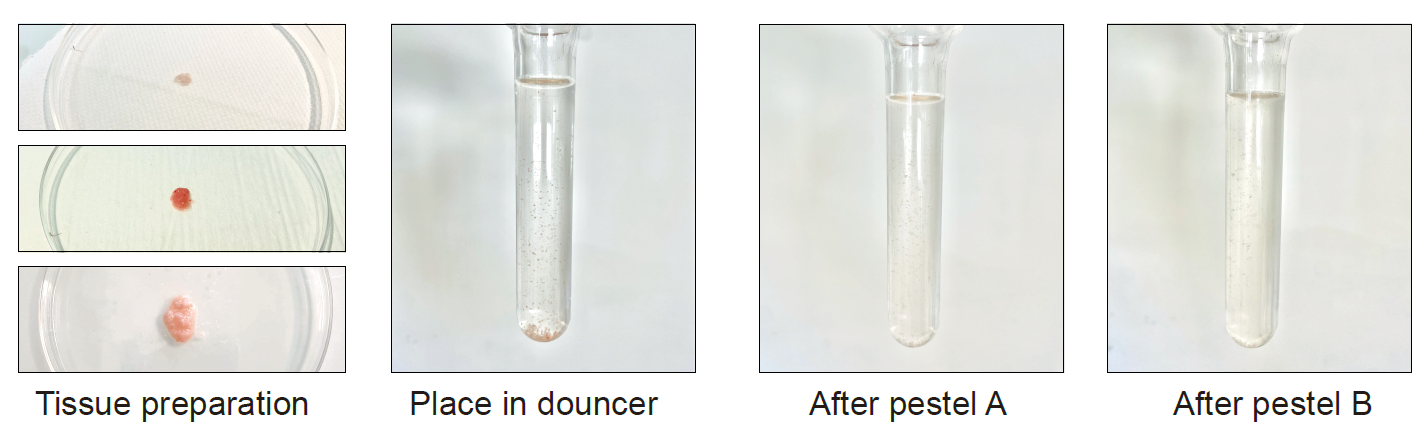
Figure 5 : Nuclei extraction from murine and human periosteum
Nuclei Sorting and counting
Nuclei sorting
Add 1 μl of SYTOX™ AADvanced™ in the solution to label nuclei.
Note: we also tested our protocol using DAPI staining with similar results.
Sort up to 150.000 nuclei (Sytox AADvanced+ nuclei) using the appropriate gating strategy (Figure X). Collect in n 1.5 ml Eppendorf containing 0.75 ml of nuclei buffer.
Note: Prior to snRNAseq experiment, cell sorting should be settled with appropriate controls including unstained nuclei.
Centrifuge the suspension of sorted nuclei at 500 × g for 5 min at 4°C. Carefully remove the supernatant without disturbing the pellet.
Critical : depending on the number of sorted nuclei, the pellet may not be visible. Proceed carefully to the supernatant removal to avoid eliminating the nuclei.
Resuspend in 50 μl of nuclei buffer.
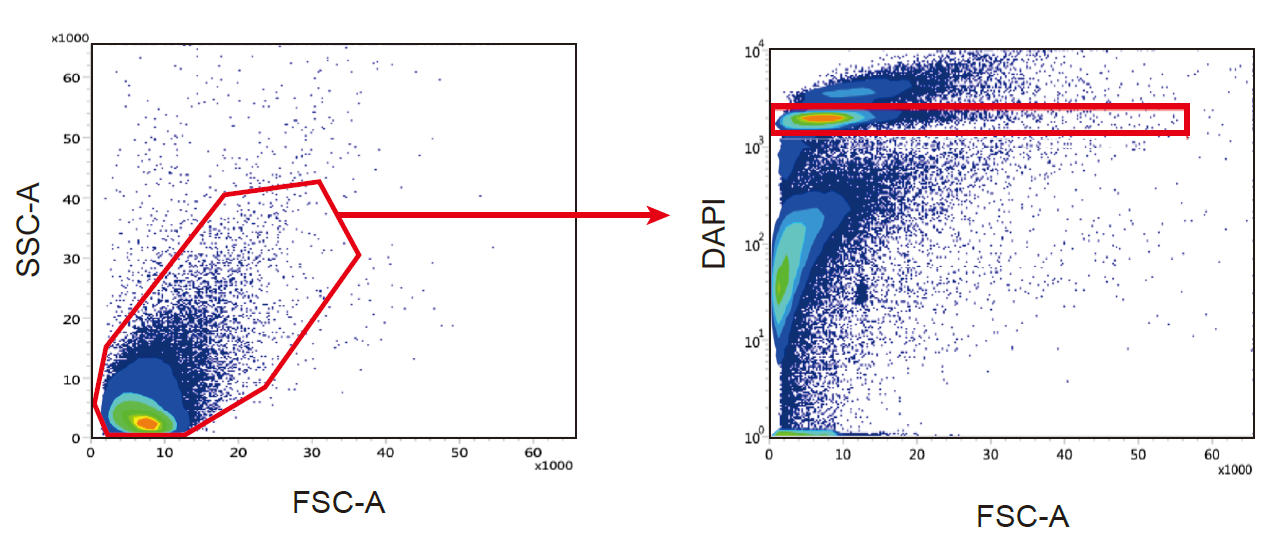
Figure 6 : Sorting strategy to obtain a nuclei suspension without debris
Nuclei counting
Mix 5 μL of the nuclei suspension with 10 μL of DAPI solution in a 0,2 mL tube.
Note : Adapt the dilution to obtain an optimal number of nuclei for counting. Too much or not enough nuclei during counting could lead to errors.
Transfer 10 μL of the stained nucleus suspension to a Kova slide.
Under a fluorescence microscope, check the quality of the nuclei suspension. Count the number of nuclei to obtain the concentration of the nucleus suspension.
Note: We recommend using both brightfield and fluorescence microscopy to check the nuclei suspension.
Critical : It is crucial to assess the quality of the nuclei suspension before proceeding to the next steps of the experiments. The suspension should only contains nuclei and no debris. The shape of the nuclei reflects the quality of the nuclei. If the suspension contains debris or a high percentage of low quality nuclei, we recommend not to use the suspension for further analysis.

Figure 7 : Nuclei counting and quality check. Nuclei should have a clean round shape. Nuclei with abnormal shape are considered low quality.
Proceed immediately to loading for snRNAseq following the manufacturer’s instructions.
Note: it is recommended to use a nuclei solution of 700–1,200 nuclei per microliter.
Validation of Protocol
This protocol was adapted from previously published protocols (G Martelotto, 2019; Santos et al., 2021), used in other studies and on various tissues. We have generated more than 10 murine samples and 6 human samples using this protocol, which were published in (Hachemi et al., 2024; Perrin et al., 2023, 2024). Datasets from the murine uninjured periosteum and fracture callus are available at the following link: https://cells.ucsc.edu/?ds=fracture-repair-atlas.
Troubleshooting
Poor data quality after snRNAseq might be due to a low quality nuclei preparation. To improve sample quality, we recommend to:
Work in a RNAse-free environment. All solutions used for the protocol must be RNAse-free. All tools used for this protocol must be sterile and cleaned to eliminate RNAse.
Reduce experimental time. Time is crucial to preserve RNA quality. Optimize all steps of nuclei extraction, sorting and process immediately to snRNAseq after nuclei extraction.
Ensure that the nuclei counting is accurate and that the suspension does not contain debris.
If working with frozen human periosteum, proper snap freezing and cryopreservation is required to obtain high quality nuclei and RNA. Reduce the time between tissue collection and freezing. To ensure the RNA quality, perform RNA extraction and check RNA integrity (RIN) from a sample from the same batch. Sample with a RIN lower than 6.5 will lead to poor results.
Acknowledgments
This work was supported by ANR-21-CE18-007-01, NIAMS R01 AR081671 and US Department of the Army NF220019 to C.C. S. Perrin, M. Ethel and C. Goachet were supported by a PhD fellowship from Paris Cité University, Univ Paris-Est Creteil and Fondation pour la Recherche Médicale respectively. This protocol was first described in Perrin et al. (2024) and in Perrin et al. (2024).
Competing interests
Authors declare no competing interest.
Ethical considerations
All procedures involving animals were approved by Paris Est Creteil University Ethical Committee (agreement #19295-2019052015468705). Sample collection from patients was approved by the Ethics Committee CPP-IDF-2 (#ID-RCB/EUDRACT: 2014- A01420-47; IMNIS2014-03).
References
Ding, J., Adiconis, X., Simmons, S. K., Kowalczyk, M. S., Hession, C. C., Marjanovic, N. D., Hughes, T. K., Wadsworth, M. H., Burks, T., Nguyen, L. T., Kwon, J. Y. H., Barak, B., Ge, W., Kedaigle, A. J., Carroll, S., Li, S., Hacohen, N., Rozenblatt-Rosen, O., Shalek, A. K., … Levin, J. Z. (2020). Systematic comparison of single-cell and single-nucleus RNA-sequencing methods. Nature Biotechnology, 38(6), Article 6. https://doi.org/10.1038/s41587-020-0465-8
Duchamp de Lageneste, O., Julien, A., Abou-Khalil, R., Frangi, G., Carvalho, C., Cagnard, N., Cordier, C., Conway, S. J., & Colnot, C. (2018). Periosteum contains skeletal stem cells with high bone regenerative potential controlled by Periostin. Nature Communications, 9(1), Article 1. https://doi.org/10.1038/s41467-018-03124-z
G Martelotto, L. (2019). ‘Frankenstein’ protocol for nuclei isolation from fresh and frozen tissue for snRNAseq v2 [Preprint]. https://doi.org/10.17504/protocols.io.3fkgjkw
Hachemi, Y., Perrin, S., Ethel, M., Julien, A., Vettese, J., Geisler, B., Göritz, C., & Colnot, C. (2024). Multimodal analyses of immune cells during bone repair identify macrophages as a therapeutic target in musculoskeletal trauma. https://doi.org/10.1101/2024.04.29.591608
Jeffery, E. C., Mann, T. L. A., Pool, J. A., Zhao, Z., & Morrison, S. J. (2022). Bone marrow and periosteal skeletal stem/progenitor cells make distinct contributions to bone maintenance and repair. Cell Stem Cell, 29(11), Article 11. https://doi.org/10.1016/j.stem.2022.10.002
Machado, L., Geara, P., Camps, J., Dos Santos, M., Teixeira-Clerc, F., Van Herck, J., Varet, H., Legendre, R., Pawlotsky, J.-M., Sampaolesi, M., Voet, T., Maire, P., Relaix, F., & Mourikis, P. (2021). Tissue damage induces a conserved stress response that initiates quiescent muscle stem cell activation. Cell Stem Cell, 28(6), Article 6. https://doi.org/10.1016/j.stem.2021.01.017
Perrin, S., & Colnot, C. (2022). Periosteal Skeletal Stem and Progenitor Cells in Bone Regeneration. Current Osteoporosis Reports. https://doi.org/10.1007/s11914-022-00737-8
Perrin, S., Ethel, M., Bretegnier, V., Goachet, C., Wotawa, C.-A., Luka, M., Coulpier, F., Masson, C., Ménager, M., & Colnot, C. (2023). Single nuclei transcriptomics reveal the differentiation trajectories of periosteal skeletal/stem progenitor cells in bone regeneration. https://doi.org/10.1101/2023.06.23.546220
Perrin, S., Protic, S., Bretegnier, V., Laurendeau, I., De Lageneste, O. D., Panara, N., Ruckebusch, O., Luka, M., Masson, C., Maillard, T., Coulpier, F., Pannier, S., Wicart, P., Hadj-Rabia, S., Radomska, K. J., Zarhrate, M., Ménager, M., Vidaud, D., Topilko, P., … Colnot, C. (2024). MEK-SHP2 inhibition prevents tibial pseudarthrosis caused by NF1 loss in Schwann cells and skeletal stem/progenitor cells. Science Translational Medicine, 16(753), eadj1597. https://doi.org/10.1126/scitranslmed.adj1597
Santos, M. D., Gioftsidi, S., Backer, S., Machado, L., Relaix, F., Maire, P., & Mourikis, P. (2021). Extraction and sequencing of single nuclei from murine skeletal muscles. STAR Protocols, 2(3), Article 3. https://doi.org/10.1016/j.xpro.2021.100694
Selewa, A., Dohn, R., Eckart, H., Lozano, S., Xie, B., Gauchat, E., Elorbany, R., Rhodes, K., Burnett, J., Gilad, Y., Pott, S., & Basu, A. (2020). Systematic Comparison of High-throughput Single-Cell and Single-Nucleus Transcriptomes during Cardiomyocyte Differentiation. Scientific Reports, 10(1), 1535. https://doi.org/10.1038/s41598-020-58327-6
Wen, F., Tang, X., Xu, L., & Qu, H. (2022). Comparison of single‑nucleus and single‑cell transcriptomes in hepatocellular carcinoma tissue. Molecular Medicine Reports, 26(5), 339. https://doi.org/10.3892/mmr.2022.12855
Wu, H., Kirita, Y., Donnelly, E. L., & Humphreys, B. D. (2019). Advantages of Single-Nucleus over Single-Cell RNA Sequencing of Adult Kidney : Rare Cell Types and Novel Cell States Revealed in Fibrosis. Journal of the American Society of Nephrology, 30(1), 23‑32. https://doi.org/10.1681/ASN.2018090912
Related files
 Perrin et al Bio-protocol 091024.pdf
Perrin et al Bio-protocol 091024.pdf - Perrin, S, Goachet, C, Ethel, M, Hachemi, Y and COLNOT, C(2024). Nuclei Isolation from Murine and Human Periosteum for Transcriptomic Analyses. Bio-protocol Preprint. bio-protocol.org/prep2731.
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link
