Advanced Search
FACS of immune cells in tumors and lymph nodes
Last updated date: Oct 18, 2022 Views: 389 Forks: 0
Abstract
Fluorescence activated cell sorting (FACS) has shown great promise in sorting and analyzing the different cell populations. Immune cells, including macrophages, T cells, B cells, dendritic cells (DCs), and natural killer (NK) cells can be infiltrated into the tumor site during the development or treatment process. This protocol provided a common method to isolate the different immune cell populations both in the tumor mass and lymph nodes.
Keywords: FACS, tumor, lymph nodes, single cell suspension
Materials and Reagents
1. RPMI 1640 medium (ThermoFisher SCIENTIFIC, catalog number: 11875119)
2. Heat-activated FBS (Sigma-Aldrich, catalog number: F9665)
3. Penicillin-Streptomycin Solution (P/S, 10,000U/mL, ThermoFisher SCIENTIFIC, catalog number: 15140122)
4. Sodium pyruvate (MilliporeSigma, catalog number: S8636-100ML)
5. L-glutamine solution (Sigma-Aldrich, catalog number: G7513)
6. DMEM (ThermoFisher SCIENTIFIC, catalog number: 10567022)
7. Hanks' Balanced Salt Solution (HBSS) (ThermoFisher SCIENTIFIC, catalog number: 14025092)
8. 60 mm dimeter cell culture dish (Corning, catalog number: 430166)
9. Surgical Scalpel (SHOPMEDVET.COM, catalog number: DS21)
10. 15 mL centrifuge tubes (MilliporeSigma, catalog number: CLS430791-500EA)
11. 50 mL centrifuge tubes (MilliporeSigma, catalog number: CLS430829-500EA)
12. FcR-blocking reagent (Miltenyi Biotec, catalog number: 130092575)
13. Red blood cell lysingbuffer hybrid-max (MilliporeSigma, catalog number: R7757)
14. 1.5 mL Eppendorf (EP) tube (MilliporeSigma, catalog number: HS4323-500EA)
15. Mouse tumor dissociation kit (MACS Miltenyi Biotec, catalog number: 130096730)
16. Strainer 70 µm (MilliporeSigma, catalog number: CLS431751-50EA) and 40 µm (MilliporeSigma, catalog number: CLS431750-50EA)
17. BSA (MilliporeSigma, catalog number: A9430)
18. DNase (MilliporeSigma, catalog number: 10104159001)
19. Antibodies (See Recipes)
20. gentleMACS C tube (MACS Miltenyi Biotec, catalog number: 130093237)
21. Cytiva HyClone™ Phosphate Buffered Saline (PBS) (Fisher scientific, catalog number: SH3025601)
22. DAPI (Sigma-AIdrich, catalog number: D9542)
23. 70% EtOH (Sigma-AIdrich, catalog number: 65350-85)
24. 5 mL FACS tubes (STELLAR SCIENTIFIC, catalog number: FSC-T9019)
Equipment
1. The gentleMACS Dissociator

2. Shaker

3. Centrifuge

4. ZE5 Flow Cytometer - High Parameter Cell Analyzer

Software
1. ZE5
2. FCS Express 7
Procedure
A. Single cell suspension preparation
1. The transgenic breast cancer bearing MMTV polyoma middle T oncoprotein (PyMT) mice (female, 15-16 weeks old with more than 10 tumors) were euthanized by carbon dioxide (CO2) inhalation following the Guiding Principles of the University Committee on Use and Care Animals at the University of Michigan.
2. Quickly dissect out the tumor mass and lymph nodes (proximal tumor) by cutting away the skin, adipose tissue, fibrous, and necrotic areas. Weight and recorded the tumor mass and lymph nodes and then place the tissues to the RPMI 1640 medium (supplemented with 2% FBS).
3. Transfer the tubes containing the tissues into a sterile hood (UV pretreated and sprayed with 70% Et-OH) and then discard the RPMI 1640 medium following by cutting the tissues into small pieces (2-4 mm).
4. Transfer the tissues into the gentleMACS C tube containing the enzyme mix (each sample: 2.35 mL DMEM, enzyme D 100 µL, enzyme R 10 µL, and enzyme A 12.5 µL).
5. Dissociate the tissues by running the gentleMACS dissociator with a tough gentleMACS program (mouse).
6. After termination the program, detach the C tube from the dissociator and incubate all samples in a shaker for 40 min at 37 ℃ (100 rpm).
7. Run the step 5 again.
8. Transfer the tissues to 50 mL tubes by filtering the 70 µm cell strainers placed onto the tubes and centrifuge the samples at 300 ×g for 5 min at 4 ℃.
9. Wash the samples by adding into 25 mL PBS into 50 mL tubes and then centrifuge the samples at 300 ×g for 5 min at 4 ℃.
10. Adding 2 mL red blood cell lysate to each tube and incubate 5 min at room temperature.
11. Terminate the lysis by adding into 10-fold PBS and then centrifuge the suspension at 300 ×g for 5 min at 4 ℃.
12. Add 200 U/mL DNase to each tube and incubate 5 min at RT.
13. Wash 300 ×g for 5 min at 4 ℃ using the PBS containing 2% BSA.
14. Discard the supernatant and HBSS buffer containing 2% BSA and count the cell numbers using a hemocytometer.
15. Quantify the cell numbers by adding HBSS buffer containing 2% BSA and the transfer the cells into 1.5 mL EP and for further antibodies staining.
B. Staining of the cell suspensions
1. Aliquot 2 million cells/sample and add DAPI (10 µg/mL) and FcR-blocking reagent to each sample and incubate in the dark for 15 min at 0 ℃.
2. Wash twice using the 0.1 M PBS containing 2% BSA and stain the cells with various antibodies (See Recipes) for 30 min at 0 ℃.
3. Wash 300 ×g for 5 min at 4 ℃ using the PBS containing 2% BSA and resuspend the cell pellet with 1mL the complete RPMI 1640 medium (supplemented with 10% FBS, 1% L-glutamine, 1% sodium pyruvate, and 1% P/S).
4. Transfer the samples into 5 mL FACS tubes (on ice) after filtering through the 40 µm strainer and then run the samples using a Bio-Rad ZE5 Flow Cytometer equipped with four lasers (405nm, 488nm, 561nm, and 640nm) and twenty-one flourescent detectors using BD FACSDiva software (BD Biosciences).
C. FACS gating for the M1 and M2 macrophages
The single live cells were isolated form the total cells in the tumor mass by the two-step singlets and DAPI negative populations (Figure 1). Then the macrophages were isolated by the CD11b and F4/80 double positive (CD11b+F4/80+) populations and then the subtype M1 phenotype macrophages and M2 phenotype macrophages were further isolated by the CD206 and CD80 markers.
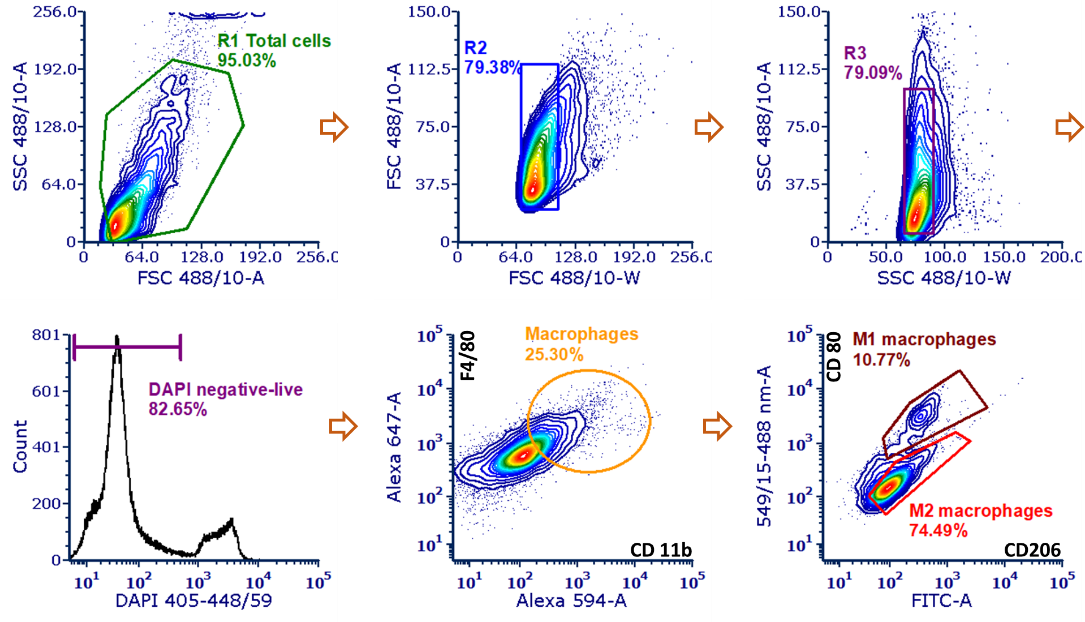
Figure 1. Representative FACS gating for M1 and M2 macrophages populations.
D. FACS gating for immune cell populations
The single live cells were isolated form the total lymphocytes cells in the tumor mass by the two-step singlets and DAPI negative populations (Figure 2). Then the total immune cells were isolated by the CD45 positive populations and then the subtype populations including macrophages (CD45+F4/80+), T cells (CD45+CD3+), B cells (CD45+CD19+), DCs (CD45+CD11c+), and NK cells (CD45+CD335+) were isolated respectively by the corresponding markers (See recipes).
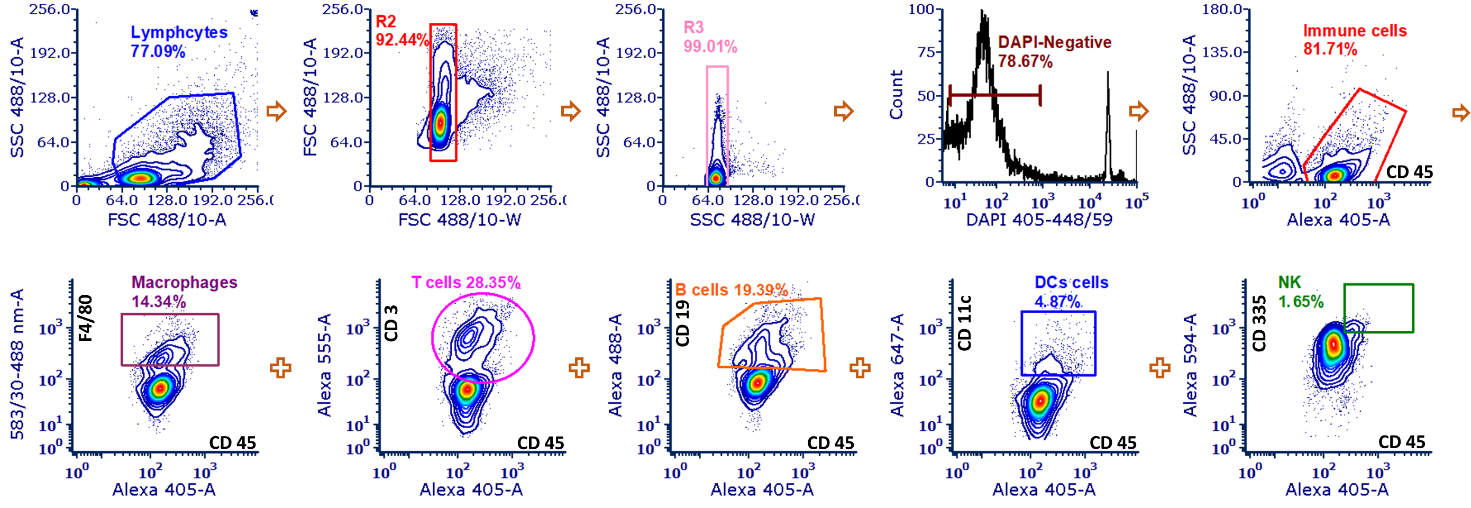
Figure 2. Representative FACS gating for variant immune cell populations.
Recipes
Antibody | Vendor | Catalog Number | Working Concentration |
| anti-F4/80-Alexa Fluor®-647 | Biolegend | 157313 | 0.25 µg per million cells in 100 µL |
| anti-CD11 b-Alexa Fluor® 594 | Biolegend | 101254 | 0.25 µg per million cells in 100 µL volume |
| anti-CD80-PE | Biolegend | 104708 | 0.5 µg per million cells in 100 µL volume |
| anti-CD206-FITC | Biolegend | 141704 | 0.125 µg per million cells in 100 µL volume |
| anti-CD45-Pacific blue | Biolegend | 157212 | 0.25 µg per million cells in 100 µL volume |
| anti-F4/80-PE | Biolegend | 123110 | 1 µg per million cells in 100 µL volume |
| anti-CD3-Spark blue 550 | Biolegend | 100260 | µg per million cells in 100 µL volume |
| anti-CD19-Alexa Fluor® 488 | Biolegend | 115521 | 0.25 µg per million cells in 100 µL volume |
| anti-CD11c-Alexa Fluor®-647 | Biolegend | 117312 | 0.25 µg per million cells in 100 µL volume |
| anti-CD335-PE/Dazzle™ 594 | Biolegend | 137630 | 0.75 µg per million cells in 100 µL volume |
Data analysis
The flow cytometry figures were analyzed through the FCS Express 7 software. Alternative software such as Flow Jo (BD Life Science) can also be used. All cell populations were isolated by the marker used which listed in the recipe tables over a minimum of three independent replicates.
Acknowledgements
This work was supported by the in part, by the internal founding of College of Pharmacy at University of Michigan. This protocol has been demonstrated to work for analyzing the immune cell populations in the tumor mass, lymph nodes, and fat pad in the BALB/C mice and PyMT mice published in Science Translational Medicine (DOI: 10.1126/scitranslmed.abl364).
Competing interests
The authors declare that they have no competing interests.
- Gao, W and Sun, D(2022). FACS of immune cells in tumors and lymph nodes. Bio-protocol Preprint. bio-protocol.org/prep1995.
- Song, Y., Bugada, L., Li, R., Hu, H., Zhang, L., Li, C., Yuan, H., Rajanayake, K. K., Truchan, N. A., Wen, F., Gao, W. and Sun, D.(2022). Albumin nanoparticle containing a PI3Kγ inhibitor and paclitaxel in combination with α-PD1 induces tumor remission of breast cancer in mice. Science Translational Medicine 14(643). DOI: 10.1126/scitranslmed.abl3649
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link
