Advanced Search
Designing and cloning of the gRNA
Last updated date: Sep 27, 2022 Views: 751 Forks: 0
Guide design:
- The guides were designed using Snap Gene and Benchling tools (https://www.benchling.com/crispr/), Benchling was used to design gRNAs for CBE while Snap Gene for ABE.
- The sequence of interest was uploaded in Benchling and then analysed for gRNA
->CRISPR->Design and Analyse guides->Parameter (guide for base editing, guide length - 20bp, Genome – homosapiens and PAM - NGG) - In case of Snap Gene the gRNAs were selected manually based on the available PAM seq in the region of interest.
- Once the gRNAs were selected, the forward and reverse sequence of the gRNA along with BsmB1 restriction sites were send for oligo synthesis.
- Forward primer consist of a gRNA without PAM (20 bp) and “CACCG”(5bp) over hang at 5’ end, while the reverse primer consist of “AAAC”(4bp) at 5’ end, a reverse complement of gRNA without PAM(20 bp) and ”C” (1bp) at 3’ end as represented below.
5’ CACCGNNNNNNNNNNNNNNNNNNNN 3’
3’ CNNNNNNNNNNNNNNNNNNNNCAAA 5’
Forward primer 5’ to 3’ - CACCGNNNNNNNNNNNNNNNNNNNN
Reverse primer 5” to 3’ - AAACNNNNNNNNNNNNNNNNNNNNC
Guide RNA cloning:
- Plasmid digestion: (Backbone)
a. Plasmids (57822(GFP)/57823(RFP)) 4ug
b. BsmB1 enzyme (NEB) 10U/ul 2ul
c. NEB buffer 3.1 10X 5ul
d. Nuclease free water make up to 50ul
e. Incubate at 55oC for 3 hours
f. Run in 1% agarose gel for 70-80 min, voltage =100V.
g. Elute using Zymo gel elution kit (Kit protocol), and store at 4oC in 50ng/ul concentration
- Oligo Annealing: using the oligos ordered for the gRNA
a. 1ul each of 100pm forward and reverse primer 2ul
b. T4 polynucleotide kinase 10U/ul (NEB) 0.5ul
c. NEB ligase buffer 10x 1ul
d. Make up to 10ul with nuclease free water
e. PCR conditions
1Cycle 1 Cycle 1 Cycle 1 Cycle
37oC 95oC 25oC 4oC
45 min 5 min 1 min ∞
Ramp 100% Ramp 100% Ramp 3%
f. Once PCR is done take 1ul from the oligo annealed product and dilute with 199ul of nuclease free water. Store the stock in -20oC and store the working in 4oC.
- Ligation:
a. Vector backbone (50ng/ul) 1ul
b. Oligo product (1:200 diluted)(Insert) 6ul
c. Ligase buffer NEB 10x 2ul
d. NEB ligase 0.5ul
e. Make up to 20ul with nuclease free water
f. Incubate in 16oC for 16 hrs for ligation.
- Transformation
a. Take B10 competent-cells (50ul) from -80oC and incubate them in ice for 15 min.
b. Add 5ul of the ligated product to the competent-cells and incubate in ice for another 15 min.
c. Now give heat shock to the cells by keeping the tubes in thermal mixer or water bath at 42oC for 45 sec.
d. Immediately, incubate the tubes in ice for 2 min.
e. After incubation, plate it in Luria bertani agar (LA) (Himedia) plate with 100µg/ml concentration of Ampicillin (SRL).
f. Spread them using a L-rod and incubate at 37oC over night
- Colony pick
a. Pick 3 colonies per plate and inoculate in 1ml of Luria bertani broth (LB) with Ampicillin in 2ml micro-centrifuge tubes each.
b. After 6 hours, take 500 µl for preparing glycerol stock and another 500µl for colony PCR
- Colony PCR:
a. Take the 500ul of inoculum, centrifuge at 4000rpm for 5min at RT.
b. Discard the supernatant and wash the pellet with 500 µl of distilled water.
c. Finally resuspend it in 500µl of distilled water for further processing.
Processed inoculum - 2ul
Forward primer (507) - 1ul (10pm) (gagggcctatttcccatgat)
Reverse primer (508) -1ul (10pm) (tggatctctgctgtccctgt)
Master mix 2x - 10ul (GoTaq® Hot Start Green Master Mix) and
Make up to 20ul with nuclease free water.
d. PCR condition is:
Initial denaturation Denaturation Annealing Extension Final extension Hold
10 min 30 sec 30 sec 45 sec 7 min
95oC 95oC 55oC 72oC 72oC 16oC
35 cycles
Product size around 550 bp
e. Take 5ul to load in 1% agarose gel to confirm the amplification.
f. Rest of the 15 µl PCR product will be used for Sanger Sequencing.
- Sequencing PCR:
Pre – clean-up
1. 20µl of Pre clean up beads (MAGBIO HighPrepTm PCR) along with 15µl of PCR product and 20µl of water were mixed thoroughly in 1.5 centrifuge tube and incubated for 5 minutes in room temperature.
2. Tubes are then placed on to the magnetic separator for 3 minutes.
3. The clear solution separated from the beads was discarded.
4. Wash the pellet twice with 70% ethanol by placing the tubes in the magnetic separator for 30sec.
5. Air dry pellet for 10 min to remove ethanol.
6. Once the pellets are dried add 20ul water and mix the pellet thoroughly
7. Let it stand in room temperature for 5 min.
8. Place the tube in magnetic separator and leave it for 5 min until the clear solution is visible, now remove the supernatant for further processing.
Sequencing PCR (BigDye Terminator v3.1 Cycle Sequencing Kit)
Reaction Components
Water - 4.5ul
Buffer - 2ul
Forward Primer - 1ul (gagggcctatttcccatgat) (10pm)
Template - 2ul (pre clean up product)
RR mix -0.5ul
Reaction conditions
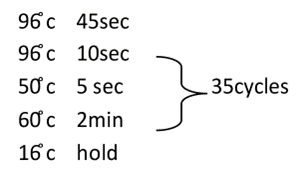
Post-clean-up
1.10ul of Post clean up beads (MAGBIO HighPrepTm DTR) along with 10ul of sequencing PCR product and 40ul of 85% ethanol were mixed thoroughly in 1.5 centrifuge and incubated for 5 minutes in room temperature.
2. Tubes are then placed on to the magnetic separator for 3 minutes.
3. The clear solution separated from the beads was discarded.
4. Wash the pellet twice with 85% ethanol by placing the tubes in the magnetic separator for 30sec.
5. Air dry pellet for 10 min to remove ethanol.
6. Once the pellets are dried add 35ul water and mix the pellet thoroughly
7. Let it stand in room temperature for 5 min.
8. Place the tube again in magnetic separator and collect the clear solution and load it in the sequencing plate.
Data processing and clone confirmation: (Finch TV or Snap Gene)
The Sanger sequencing data were analysed for the positive clones using Finch TV or Snap Gene tools. Once a positive clone is confirmed the respective glycerol stock is store and the rest discarded.
- Mohankumar, K and R, N(2022). Designing and cloning of the gRNA. Bio-protocol Preprint. bio-protocol.org/prep1968.
- Ravi, N. S., Wienert, B., Wyman, S. K., Bell, H. W., George, A., Mahalingam, G., Vu, J. T., Prasad, K., Bandlamudi, B. P., Devaraju, N., Rajendiran, V., Syedbasha, N., Pai, A. A., Nakamura, Y., Kurita, R., Narayanasamy, M., Balasubramanian, P., Thangavel, S., Marepally, S., Velayudhan, S. R., Srivastava, A., DeWitt, M. A., Crossley, M., Corn, J. E. and Mohankumar, K. M.(2022). Identification of novel HPFH-like mutations by CRISPR base editing that elevate the expression of fetal hemoglobin. eLife. DOI: 10.7554/eLife.65421
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link
