Advanced Search
Measuring resistance-infectivity (co)evolution
Last updated date: May 26, 2022 Views: 660 Forks: 0
Time-shift assay protocol: Castledine et al., (2022) eLife
To assess whether bacteria and phage had coevolved in vivo and in vitro, resistance assays were conducted where bacteria were exposed to phage populations from the same/contemporary, past, and future time points (time-shift assays) (Buckling and Rainey, 2002). If bacteria and phage coevolved, both bacteria and phage would show significant changes in resistance and infectivity, respectively, through time. Increases in resistance and infectivity would be indicative of arms race dynamics, while nondirectional changes would be indicative of fluctuating selection dynamics (Lopez Pascua et al., 2014).
In vivo
Materials:
- Round LB agar plates (x 1 per bacterial sample) – for resistance testing against ancestral phages 14-1 and PNM
- Square LB agar plates (x 1 per bacterial sample) – for resistance testing against phages isolated in vivo (12 phages per time-point, 3 time point)
- LB soft agar
- 10 µl and 100 µl pipettes and pipette tips
- Glass vials (25 mL)
- Incubator (37)
- Orbital shaker
- Grow bacterial samples overnight from days 2 and 4 of treatment in individual glass vials (37°C, shaking at 180 r.p.m.)
- Inoculate 100 µl of bacterial cultures into individual 15mL tubes containing 10 mL soft LB agar and 100 µl into individual 15mL tubes containing 5-8 mL soft LB agar
- Pour the 10mL soft agar / bacterial mix onto individual square LB agar plates and the 5-8 mL soft agar / bacterial mix onto individual round LB agar plates and leave to dry
- Spot 5 µl of individual phage isolates (n = 12 per time point (3 time points of days 2, 4 and 7 of treatment from which phage could be isolated) onto square plates). Ancestral phage, 14-1 and PNM, were spotted onto round plates of each bacterial Leave to dry (~20 mins).
- Turn plates upside down and incubate overnight (37°C)
- Bacterial isolates were scored as 1 to susceptible and 0 to resistant if at least 1 phage isolate from each time-point could infect (showing lack of bacterial growth) (Figure 1 and 2)
In vitro
In vitro isolates from days 4, 8, and 12 were tested (16 colonies per time point) against phage populations isolated from each time point (Figure 6). Each treatment replicates from each time point were defined as biological replicates, while clonal isolates are biological replicates nested within each treatment replicate.
Materials:
- Round LB agar plates
- LB soft agar
- 10 µl and 100 µl pipettes and pipette tips
- Glass vials (25 mL)
- Incubator (37°C)
- Orbital shaker
- Grow bacterial clones (n = 16 per treatment replicate and time-point) overnight from 3 time-points of treatment in individual glass vials (37°C, shaking at 180 r.p.m.)
- Inoculate 100 µl of bacterial cultures into individual 15mL tubes containing 5-8 mL soft LB agar
- Pour the 5-8 mL soft agar / bacterial mix onto individual round LB agar plates and leave to dry
- Spot 5 µl of phage isolates from each time point (isolated from the same treatment replicate as the bacteria isolated in the overlay e.g. replicate 1 phage (time-points 1-3) with replicate 1 bacteria). Leave to dry (~20 mins).
- Turn plates upside down and incubate overnight (37°C)
- Bacterial isolates were scored as 1 to susceptible and 0 to resistant if at least 1 phage isolate from each time-point could infect (showing lack of bacterial growth)
Resistance to ancestral phage: ancestral bacteria and in vitro bacteria (day 12)
Materials:
- Round LB agar plates
- LB soft agar
- 10 µl and 100 µl pipettes and pipette tips
- Glass vials (25 mL)
- Incubator (37°C)
- Orbital shaker
- Grow bacterial clones (n = 24 ancestral bacteria, n = 24 (per replicate) day 12 in vitro bacteria) overnight from 3 time-points of treatment in individual glass vials (37°C, shaking at 180 r.p.m.)
- Inoculate 100 µl of bacterial cultures into individual 15mL tubes containing 5-8 mL soft LB agar
- Pour the 5-8 mL soft agar / bacterial mix onto individual round LB agar plates and leave to dry
- Spot 5 µl of each ancestral phage onto each plate. Leave to dry (~20 mins).
- Turn plates upside down and incubate overnight (37°C)
- Bacterial isolates were scored as 1 to susceptible and 0 to resistant to each phage (14-1 and PNM) if lack of growth or growth shown respectively.
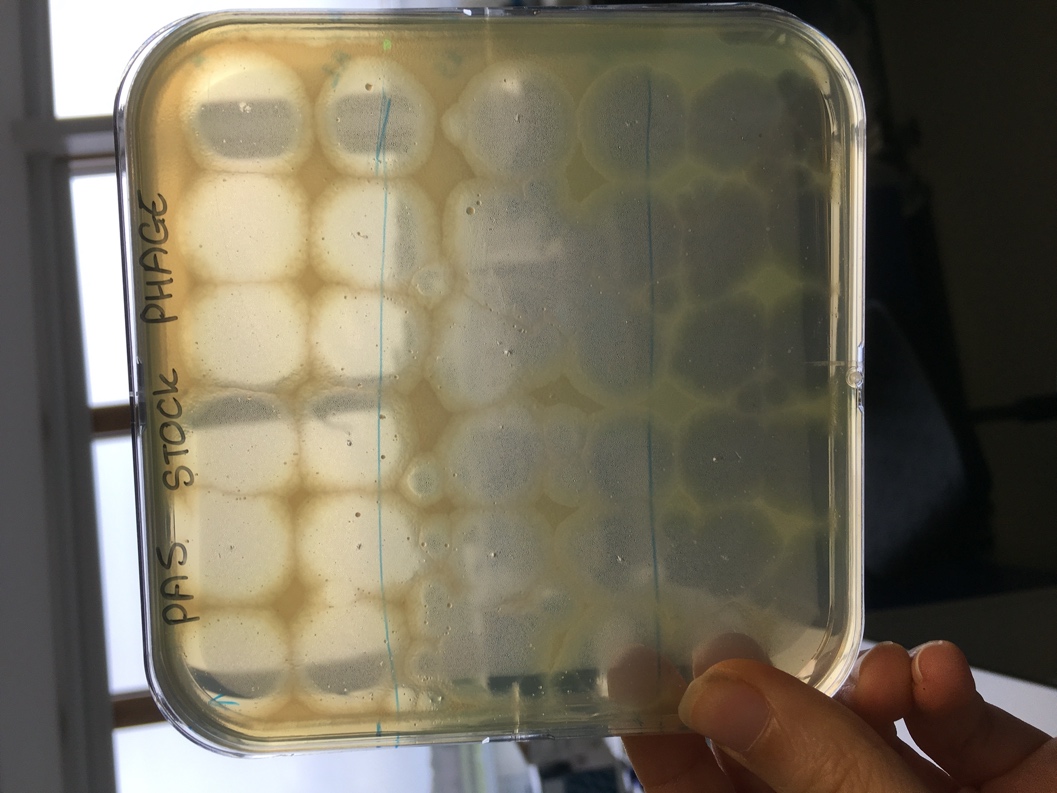
Figure 1. An example of a square LB agar plate with LB soft overlay of Pseudomonas aeruginosa. Cleared plaques indicate successful phage infection (susceptibility) from different phage clones.

Figure 1. An example of one replicate in which an in vivo bacterial isolate from day 2 of treatment was exposed to phage from days 2 (52), 4 (53) and 7 (54) of treatment. Cleared plaques at 53 and 54 indicate one phage isolate could infect from each preparation, indicating susceptibility at that time-point. In this example, no plaques at 52 (top section) indicate no successful infections (complete resistance) to phage from its own time-point.
- Castledine, M(2022). Measuring resistance-infectivity (co)evolution. Bio-protocol Preprint. bio-protocol.org/prep1684.
- Castledine, M., Padfield, D., Sierocinski, P., Soria Pascual, J., Hughes, A., Mäkinen, L., Friman, V., Pirnay, J., Merabishvili, M., de Vos, D. and Buckling, A.(2022). Parallel evolution of Pseudomonas aeruginosa phage resistance and virulence loss in response to phage treatment in vivo and in vitro. eLife. DOI: 10.7554/eLife.73679
Category
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link
