Advanced Search
Generation of erythroid-specific TLR9-deficient mice
Last updated date: Mar 17, 2022 Views: 763 Forks: 0
Generation of erythroid-specific TLR9-deficient mice
Abstract
We have recently reported that red blood cells (RBCs) express the nucleic acid sensor and pattern recognition receptor TLR9. To investigate the in vivo function of TLR9 on erythrocytes, we generated erythroid-specific TLR9-deficient (Erytlr9-/-) mice using the Cre-loxP system. Our protocol presented here includes the procedure for breeding Erytlr9-/- mice, genotyping of the mice via PCR, and confirming the phenotype using flow cytometry and immunofluorescence staining.
Background
Red blood cell immune function is often neglected despite growing evidence of their immunomodulatory capabilities. 1 Because numerous other cell types express immune genes robustly; studies using global genetic knockout or bone marrow chimera are unsuitable for examining RBC-specific immunomodulatory properties in vivo, whereas exchange transfusion experiments can be confounded by the physiological changes incurred due to transfusion. Although RBCs are abundant in the mammalian body, investigating physiological manifestations due to genetic variations in RBCs remain a challenge in vitro. Because mature RBCs lack nuclei, direct genetic modification of RBCs requires manipulation of its progenitor cells and subsequent differentiation parameters, which may not resemble the complex erythropoietic processes during altered inflammatory states such as stress erythropoiesis. Therefore, an erythroid-specific genetic modified mouse is a powerful tool for understanding the specific functions of immune molecules on RBCs in a physiologically relevant context.
We recently reported that RBCs bind CpG-containing DNA through TLR9, and CpG-carrying RBCs trigger inflammation and undergo accelerated erythrophagocytosis. 2 To understand the role of RBC-TLR9 in vivo and its role in sepsis, we generated erythroid-specific TLR9-deficient (Erytlr9-/-) mice using the Cre-loxP system, by crossing mice with Cre expression driven by the erythropoietin receptor promoter3 and mice with Tlr9 flanked by loxP sites. Using Erytlr9-/- mice, we further demonstrated that TLR9 deficiency in RBCs dampens the inflammation triggered by CpG or sepsis. 2 Therefore, mice possessing erythroid-specific genetic modifications can be used to investigate protein function on mature RBCS. Here, we detail our protocols for generating erythroid-specific TLR9-deficient mice and the subsequent verification of phenotype.
Materials and Reagents:
- Animals
- ErGFP Cre mouse (EpoR Cre, provided by Dr. Ursulla Klingmueller, also available at Jackson Laboratory, strain #035702)
- TLR9 conditional knockout mouse (The European Mouse Mutant Archive, ID: 05497)
- Genotyping Animals
- Alcohol wipe (Cardinal Health 200, Inc., catalog#6818)
- Alkaline lysis reagent (see Recipes)
- Neutralization buffer (see Recipes)
- Nuclease-free water (Invitrogen, catalog#AM9932)
- GoTaq® G2 Green Master Mix (Promega, catalog#M7122)
- Primer pair for EpoRCre (see Table I)
- Primer pair for TLR9 flx (see Table I)
- Tris-borate-EDTA buffer (TBE buffer, Bio-Rad, catalog#1610733)
- 1% agarose gel with 0.5 μg/mL ethidium bromide
- UltraPure Agarose (Invitrogen, catalog#16500-500)
- Ethidium bromide (Sigma-Aldrich, catalog#E1510)
- Isolation of RBCs
- Leibovitz L-15 media (MACS buffer, Gibco, catalog#11415-064)
- Anti-Ter-119 MicroBeads, mouse (Miltenyi Biotec Inc., catalog#130-049-901)
- MACS LS columns (Miltenyi Biotec Inc., catalog#130-042-401)
- Flow cytometry and immunofluorescence staining
- Phosphate buffer saline (PBS, Corning, catalog#21-031-CM)
- 8% Glutaraldehyde (Electron Microscopy Science, catalog#16019, 4°C storage)
- Permeabilization buffer (see Recipes)
- Wash buffer (see Recipes)
- FACS buffer (see Recipes)
- Sodium azide (Fisher Scientific, catalog#S227I-25)
- Normal goat serum (Cell Signaling Technology, catalog#5425S)
- Fetal bovine serum (FBS, VWR, catalog#1500-500)
- Bovine serum Albumin (BSA, R&D Systems, catalog#DY995)
- Triton X-100 (ThermoFisher Scientific, catalog#28314)
- Tween20 (ThermoFisher Scientific, catalog#28320)
- Blocking buffer (see Recipes)
- Antibody diluent (see Recipes)
- Rat anti-TER119 antibody (clone TER119, Biolegend, catalog#116211)
- Mouse anti-TLR9 antibody FITC-conjugated (clone 5G5, Abcam, catalog#ab58864)
- Rabbit anti-TLR9 antibody (Abcam, catalog# ab37154)
- Goat anti-rat IgG antibody, AlexaFluor488-conjugated (Jackson ImmunoResearch, catalog#112-545-167)
- Goat anti-rabbit IgG antibody, AlexaFluor647-conjugated (Jackson ImmunoResearch, catalog#111-605-144)
- Coverslip (Electron Microscopy Science, catalog# 72195-15 )
- Superfrost plus glass slide (Fisher Scientific, catalog#12-550-15)
Equipment:
- Surgical scissors (Fine Science Tools, catalog#14059-11)
- Mouse ear tag plier (National Band & Tag Company, catalog#1005S1)
- Ear tags (National Band & Tag Company, catalog#10051)
- Thermocycler (ThermoFisher Scientific, catalog#A37834)
- Gel imager, Amersham Imager 680 (General Electric, catalog#29270769)
- Nutator (Fisher Scientific, catalog#05-250-213)
- Flow Cytometer (BDFortessa)
- FlowJo Software
- Nikon 2A fluorescent microscope
Procedure:
- Animals:
- Breeding Erythroid-specific TLR9-deficient mice (Erytlr9-/-) mice
- To generate Erytlr9-/-, cross breed EpoRCre mice with TLR9 flx mice for F1 heterozygous generation.
- Genotype the animals (see below).
- Cross breed the F1 generation with each other.
- Erytlr9-/- are mice expressing EpoR Cre and homozygous for TLR9 flx allele.
- Weaning and Tagging
- Wean the offspring mice from the breeding cages at 3 weeks old.
- Place an ear tag in the plier.
- Prepare an Eppendorf tube that has the corresponding ear tag number.
- Disinfect surgical scissors with alcohol wipe.
- Scruff and restrain the mouse, and then place the tag onto the inner cartilage of ear and close the plier to tag it.
- Cut a piece of mouse tail no longer than 2 mm and place into the pre-labeled tube.
- Apply pressure to mouse tail to stop any bleeding if necessary.
- Store tail snip at -20°C until needed.
- Breeding Erythroid-specific TLR9-deficient mice (Erytlr9-/-) mice
- Genotyping
- Obtain tube with tail snip.
- Add 75 μL of Alkaline Lysis Reagent.
- Heat at 95°C for 30minutes.
- Cool at 4°C for 5-10 minutes.
- Centrifuge briefly to collect liquid.
- Add 75 μL Neutralization Buffer and mix well.
- Extracted DNA can be stored at -20°C until usage.
- Dilute primers to 10 μM in nuclease-free water.
- Prepare PCR reaction mix by combining 10 μL GoTaq Green Master Mix, 0.2 μL of each of the two primers, 8.6 μL of nuclease-free water perreaction.
- Add 19 μL reaction mix to each PCR tube.
- Add 1 μL extracted DNA sample to the PCR tubes with reaction mix.
- Mix well and centrifuge briefly to collect volume to the bottom of the tube.
- Perform PCR reactions in a thermocycler according table II.
- Store PCR tubes in 4°C until needed.
- PCR product should be analyzed within 24 hour.
- Resolve PCR product with agarose gel electrophoresis.
- Use 1% agarose gel made in 1x TBE buffer.
- Load 10 μL of either the DNA ladder or PCR product to eachwell.
- Electrophoresis for 1 hour at 100 V.
- PCR products were visualized on the Amersham Imager 680 under UV illumination.
- Interpret the results according to table 1, a representative result is shown in Figure 1
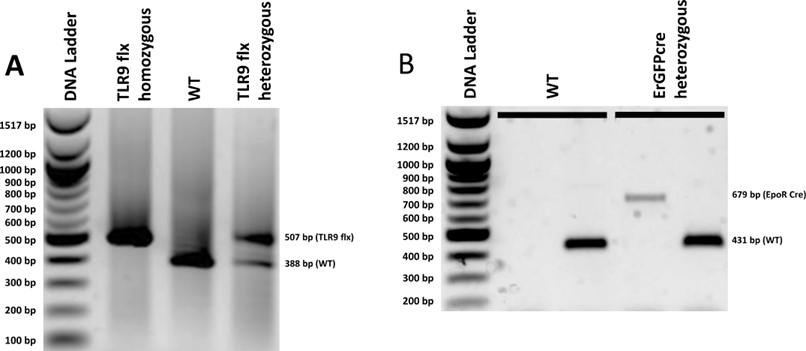
Figure 1. Genotyping of Erytlr9-/- mice (A) Agarose gel analysis of PCR products amplified with TLR9 flx (A),and EpoR Cre AC and EpoR Cre BC (B). In (B), the left well for each sample represents the PCR products amplified with EpoR Cre AC, and the right well for each sample represents the PCR products amplified with EpoR Cre BC. The two samples in (B) include a WT as well as a ErGFPcre heterozygote. Gel electrophoresis was performed on 1% agarose gels with 0.5 ug/mL ethidium bromide to stain. This gel contains a DNA ladder with the corresponding base pair size of each band on the left side and the expected sample band positions on the right side.
C. Isolation of RBCs
- Obtaining Blood Samples
- Anesthetize mouse with an 80 mg ketamine/10 mg xylazine/kg of mouse mixture.
- Obtain whole blood via cardiac puncture.
- Store blood in EDTA tube on ice until RBC purification.
- Euthanize mouse via cervical dislocation.
- RBC Purification
- Add 30 μL of whole blood to 70 μL of Leibowitz media (MACS buffer) in a 1.5 mL Eppendorf tube. Ensure that reagents are kept cold.
- Add 15 μL of Anti-TER119 beads to the Eppendorf tube.
- Mix well and incubate for 15 minutes on a nutator at 4°C.
- Wash cells by adding 1mL of MACS buffer. Centrifuge 300 g for 5 minutes, and then remove the supernatant.
- Resuspend cells in 500 μL of MACS buffer.
- Place LS column into the magnetic field of a MACS separator. Save the associated plunger for later use.
- Prepare column by rinsing it with 3 mL of MACS buffer.
- Add cell suspension into the column and let the suspension run through the column before proceeding to washes.
- Wash the column with 3 mL of MACS buffer twice.
- Remove column from the separator and place it in a 15 mL conical tube.
- Add 5 mL of MACS buffer into the column and use the plunger to flush out the column.
- Centrifuge for 5 minutes at 800 g with a slow deceleration setting of 6.
- Remove the supernatant and wash once with 5 mL of PBS.
- Repeat centrifugation to pellet cells.
- Remove supernatant and resuspend cells in 1 mL of PBS.
- Take 10 μL of cells and perform a serial dilution. Count the number of cells on a hemacytometer.
- Resuspend in PBS to achieve 108 cells/mL.
D. Flow Cytometry staining for fresh RBCs
- Dilute freshly isolated RBCs to 107 cells/mL.
- Add 100 μL (106 cells) to polypropylene tubes for each staining reaction.
- Centrifuge at 800 g for 5 minutes with a deceleration of 6.
- Remove supernatant.
- Add 5 μg of FITC-conjugated 5G5 antibody or isotype diluted in FACS buffer to cells.
- Add FACS buffer to one tube as unstained.
- Incubate at room temperature for 45 minute under gentle shaking conditions.
- Wash cells with 1 mL of FACS buffer twice.
- Centrifuge at 800 g for 5 minutes with a deceleration of 6.
- Remove supernatant.
- Resuspend cells in 400 μL FACS buffer.
- Analyze fluorescence with a flow cytometer.
E. Fixation of Cells
- Prepare 0.1% glutaraldehyde in PBS.
- Resuspend RBCs to 107 cells/mL in PBS.
- Add an equal volume of the 0.1% glutaraldehyde to achieve cell suspension in a final concentration of 0.05% glutaraldehyde.
- Incubate the tube at room temperature in the dark for 10 minutes.
- Wash twice with FACS buffer.
- Add an equal volume of FACS buffer to the tube.
- Centrifuge at 800 g for 5 minutes.
- Discard supernatant and repeat wash once more.
- Resuspend fixed cells in 1 mL FACS buffer supplemented with 0.05% NaN3
- Store at 4°C untilneeded.
F. ImmunofluorescenceStaining
- Obtain 106 fixed RBCs for each staining reaction.
- Wash cells once with FACS buffer.
- Resuspend cells in 200 μL of 0.1% TritonX-100 diluted in FACS buffer.
- Incubate for 15 minutes at room temperature.
- Add 800 μL FACS buffer.
- Centrifuge at 800 g for 5 minutes and discard supernatant.
- Wash with 1 mL FACS buffer twice.
- Block permeabilized cells in blocking buffer for 1 hour at room temperature.
- Dilute primary antibodies (5 μg/mL ab37154 and 1μg/mL TER-119, 100 μL per stain) in antibody diluent and place on ice until usage.
- After blocking, pellet cells at 800 g for 5 minutes.
- Discard supernatant and resuspend cells in 100 μL antibody cocktail.
- Incubate overnight at 4°C under gentle agitation.
- Wash cells with 1 mL PBST for three times, centrifuging at 800 g for 5 minutes in between.
- Dilute secondary antibodies (1:400 for both antibodies, 100 μL per stain).
- After washes, resuspend cells in 100 μL of diluted secondary antibodies.
- Incubate at room temperate in the dark for 1 hour.
- Wash three times in PBST as above.
- Resuspend cells in no more than 10 μL PBS.
- Add 5 μL Fluoromount-G onto a glass slide and let air dry.
- Add stained cells and mix gently to avoid bubble.
- Slowly lower a coverslip onto the mounted sample.
- Blot off excess liquid and seal sample with nail polish.
- Store slides at 4°C under dark until imaging.
Recipes
- Alkaline lysis reagent: 25 mM NaOH with 0.2 mM EDTA in nuclease-free water
- Neutralization buffer: 4 mM Tris-HCl in nuclease-free water
- Permeabilization buffer: PBS with 0.1% Triton X-100
- PBST: PBS with 0.05% tween20
- FACS buffer: PBS with 2% fetal bovine serum
- Blocking buffer: PBST with 1% BSA and 5% normal goat serum
- Antibody diluent: PBST with 1% BSA Table I. Primers and expected results for genotyping
| Gene Target | Primer 1 | Primer 2 | Expected Band(s) |
| TLR9 flx | TLR9 1514-27-F: 5’-CGG TTA ATG GTA GCA CTT GG-3’ | TLR9 1514-28-R: 5’-GCT TTT GCT CAG AAC ACA ACC-3’ | WT = 388 bp; TLR9 flx = 507 bp |
EpoR Cre | A 5’-GTG TGG CTG CCC CTT CTG CAA-3’ | C Common: 5’-CAG GAA TTC AAG CTC AAC CTC A-3’ | EpoR Cre = 679 bp |
| B 5’ GGC AGC CTG GGC ACC TTC AC-3’ | C Common: 5’-CAG GAA TTC AAG CTC AAC CTC A-3’ | WT = 431 bp |
Table II. Thermocycling programs for PCR
| TLR9 floxed | EpoR Cre | ||||
| Denature | 95 °C | 5 min | 95 °C | 5 min | |
Amplification | Melting | 94 °C | 30 sec | 94 °C | 20 sec |
| Annealing | 60 °C | 45 sec | 60 °C | 20 sec | |
| Polymerization | 72 °C | 45 sec | 72 °C | 45 sec | |
| Extension | 72 °C | 10 min | 72 °C | 7 min | |
| Cooling | 12 °C | hold | 4 °C | hold | |
36 cycles TLR9 flx; 39 cycles EpoR Cre
Acknowledgements The research was supported by the following grants: NIH grant R01 HL126788 to N.S.M.
Competing Interests The authors declare no conflicts of interest.
- Anderson HL, Brodsky IE, Mangalmurti NS. The Evolving Erythrocyte: Red Blood Cells as Modulators of Innate Immunity. The Journal of Immunology. 2018;201(5):1343-1351.
- Lam LKM, Murphy S, Kokkinaki D, et al. DNA binding to TLR9 expressed by red blood cells promotes innate immune activation and anemia. Science Translational Medicine. 2021;13(616):eabj1008.
- Heinrich AC, Pelanda R, Klingmuller U. A mouse model for visualization and conditional mutations in the erythroid lineage. Blood.2004;104(3):659-666.
- Lam, L M, Eckart, K and Mangalmurti, N(2022). Generation of erythroid-specific TLR9-deficient mice. Bio-protocol Preprint. bio-protocol.org/prep1588.
- Lam, L. K. M., Murphy, S., Kokkinaki, D., Venosa, A., Sherrill-Mix, S., Casu, C., Rivella, S., Weiner, A., Park, J., Shin, S., Vaughan, A. E., Hahn, B. H., John, A. R. O., Meyer, N. J., Hunter, C. A., Worthen, G. S. and Mangalmurti, N. S.(2021). DNA binding to TLR9 expressed by red blood cells promotes innate immune activation and anemia. Science Translational Medicine 13(616). DOI: 10.1126/scitranslmed.abj1008
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link
