Advanced Search
Computational-driven epitope verification and affinity maturation of TLR4-targeting antibodies
Last updated date: Feb 2, 2022 Views: 750 Forks: 0
Computational-driven epitope verification and affinity maturation of TLR4-targeting antibodies
Bilal Ahmad 1, Maria Batool 1,2, Moon Suk Kim1 and Sangdun Choi 1,2,*
1Department of Molecular Science and Technology, Ajou University, Suwon, 16499, Korea; bilalpharma77@gmail.com (B.A.); mariabatool.28@gmail.com (M.B.); moonskim@ajou.ac.kr (M.S.K.)
2S&K Therapeutics, Woncheon Hall 135, Ajou University, Suwon, 16499, Korea
*Correspondence: ; Fax: +82-31-219-1615; Tel: +82-31-219-2600
4.3. Epitope prediction and CDR annotations
The RCSB PDB database was used to extract the 3D coordinates of TLR4 (PDB ID: 3FXI) and TLR4-specific monoclonal antibodies (mAbs) Hu 15C1 (PDB ID: 4R7D) and C2E3 (PDB ID: 4R7N).The TLR4–MD2 complex was used to create a monomeric TLR4 structure, which was then optimized in the 2019 molecular operating environment (MOE). After removing the water of crystallization, hydrogen atoms and partial charges were added, incomplete and broken side chains were redesigned, and hybridization events were regulated in MOE at default structure preparation conditions. To predict the conformational epitope, the prepared TLR4 and mAb structure was subjected to Epipred tool of the SAbPred [1,2]web server for the prediction of conformational epitopes. Antigen epitopes are classified according to the combined conformational matching of antibody-antigen structures through geometric fitting and knowledge-based asymmetric antibody-antigen score. An epitope's score is determined by:
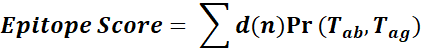
where Tab and Tag are the amino acid types of the antibody and antigen residues that correspond to node n, respectively. The Ramachandran plot was used to validate the stereochemistry and geometry of chosen epitopes using the MOE's inbuilt functionality. The residues forming the conformational epitope can be visualized in MOE by selecting the b-factor the epitopic residues will be highlighted.
Standardized numbering methods are required to precisely define complementary determining regions (CDR), frameworks, and light- and heavy-chain residues that affect the binding affinity and/or specificity of the antibody-antigen interaction, which is a prerequisite for antibody humanization. The most widely used schemes, which are now universally acknowledged by immunologists, were developed by Kabat and Chothia. The other antibody numbering schemes are Martin [3], Gelfand [4-6], IMGT [7], and Honeger (Aho) [8]. Chothia and Lesk's [9,10] numbering technique was used to annotate CDRs of antibodies acquired from the PDB database. CDRs of mAbs were used as ligand sites in docking and simulations instead of the whole Fab region.
References
1. Dunbar, J., Krawczyk, K., Leem, J., Marks, C., Nowak, J., Regep, C., et al. (2016). SAbPred: a structure-based antibody prediction server. Nucleic Acids Res 44(W1), W474-478. doi: 10.1093/nar/gkw361.
2. Krawczyk, K., Liu, X., Baker, T., Shi, J., and Deane, C.M. (2014). Improving B-cell epitope prediction and its application to global antibody-antigen docking. Bioinformatics 30(16), 2288-2294. doi: 10.1093/bioinformatics/btu190.
3. Allcorn, L.C., and Martin, A.C. (2002). SACS--self-maintaining database of antibody crystal structure information. Bioinformatics 18(1), 175-181. doi: 10.1093/bioinformatics/18.1.175.
4. Gelfand, I.M., and Kister, A.E. (1995). Analysis of the relation between the sequence and secondary and three-dimensional structures of immunoglobulin molecules. Proc Natl Acad Sci U S A 92(24), 10884-10888. doi: 10.1073/pnas.92.24.10884.
5. Stoyanov, O., Kister, A., Gelfand, I., Kulikowski, C., and Chothia, C. (2000). Geometric invariant core for the CL and CH1 domains of immunoglobulin molecules. J Comput Biol 7(5), 673-684. doi: 10.1089/106652701446143.
6. Gelfand, I.M., Kister, A.E., and Leshchiner, D. (1996). The invariant system of coordinates of antibody molecules: prediction of the "standard" C alpha framework of VL and VH domains. Proc Natl Acad Sci U S A 93(8), 3675-3678. doi: 10.1073/pnas.93.8.3675.
7. Warr, G.W., Clem, L.W., and Soderhall, K. (2003). The international ImMunoGeneTics Database IMGT. Dev Comp Immunol 27(1), 1. doi: 10.1016/s0145-305x(02)00094-0.
8. Honegger, A., and Pluckthun, A. (2001). Yet another numbering scheme for immunoglobulin variable domains: an automatic modeling and analysis tool. J Mol Biol 309(3), 657-670. doi: 10.1006/jmbi.2001.4662.
9. Chothia, C., and Lesk, A.M. (1987). Canonical structures for the hypervariable regions of immunoglobulins. J Mol Biol 196(4), 901-917. doi: 10.1016/0022-2836(87)90412-8.
10. Murzin, A.G., Brenner, S.E., Hubbard, T., and Chothia, C. (1995). SCOP: a structural classification of proteins database for the investigation of sequences and structures. J Mol Biol 247(4), 536-540. doi: 10.1006/jmbi.1995.0159.
- Ahmad, B(2022). Computational-driven epitope verification and affinity maturation of TLR4-targeting antibodies. Bio-protocol Preprint. bio-protocol.org/prep1522.
- Ahmad, B., Batool, M., Kim, M. and Choi, S.(2021). Computational-Driven Epitope Verification and Affinity Maturation of TLR4-Targeting Antibodies. International Journal of Molecular Sciences 22(11). DOI: 10.3390/ijms22115989
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link
