Advanced Search
Quantitation of the Monoclonal Antibody Conjugated to Nanosomes
Last updated date: Jun 17, 2021 Views: 1056 Forks: 0
Title: Quantitation of the Monoclonal Antibody Conjugated to Nanosomes
Authors: Fazal Ur Rehman Bhatti1,2, Su-Jin Heo4, and Hongsik Cho1,2,3 *
Authors’ affiliation(s) and address
1Department of Orthopaedic Surgery and Biomedical Engineering, The University of Tennessee Health Science Center, Memphis, TN, USA
2Memphis VA Medical Center, Memphis, TN, USA
3Campbell Clinic, Memphis, TN, USA
4Department of Orthopaedic Surgery, University of Pennsylvania, Philadelphia, PA, USA
*To whom correspondence should be addressed:
Hongsik Cho, PhD, MBA
Department of Orthopaedic Surgery and Biomedical Engineering,
University of Tennessee Health Science Center
Research 151, VAMC, 1030 Jefferson Ave, Memphis TN 38104 USA
Phone: 1-901-523-8990 (Ext: 6456)
Fax: 1-901-577-7273
E-mail: hcho4@uthsc.edu
Abstract:
The nano-sized liposomes known as ‘nanosomes’ are used to deliver drug of interest due to their numerous advantages, such as prevention of drug degradation, controlled drug release, enhanced pharmacokinetics, systemic drug delivery with minimal side-effects, a longer period of circulation, and target delivery to the diseased tissues. The selective delivery of the drug of interest to the target tissue via nanosomes can be enhanced by attaching surface ligands, such as a monoclonal antibody (mAb) to the nanosomes. The current methods that are used to quantify mAb concentration include immunoassays and liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS). However, these methods are laborious, time-consuming, and require highly skilled labor to perform these techniques. Therefore, here we provide a simple, easy, and time-effective method for the quantification of mAb attached to the targeted nanosomes. Briefly, targeted nanosomes are lyzed by 1% Triton X-100, centrifuged to remove excess lipids, and read by the NanoDrop instrument. The results can be verified against known antibody standards. Thus, we present here a robust and reliable method for the quantification of mAb attached to nanosomes that will save time and labor for other researchers in the field.
Keywords: Liposomes, antibody, targeted-drug delivery, ligand-targeting, quantification
1.Background
Liposomes have been utilized for therapeutic purpose since their discovery in 1960s, owing to their flexible structure and practicality in function [1]. The beneficial aspects of using liposomes as drug delivery system (DDS) include transport and protection of diverse type of drugs, biocompatibility, biodegradation, various forms of desirable morphology, ease of manipulation in chemical composition, low level of toxicity, non-immunogenic nature and reduced cost [2, 3]. Although liposomes can be modified in a variety of ways, however, active targeting has gained considerable attention. Active targeting enhances targeting capability, selectivity, cellular internalization, prolongs exposure, improves therapeutic index and reduces the systemic side effects [4, 5]. One of the prominent examples of active targeting of liposomes include ligand-targeted liposomes (LTLs) that carry a molecule of interest, such as a monoclonal antibody, on their surface that enables the liposomes to bind to the target tissue and release the drug of interest at a specific site [6]. This approach has been widely used in cancer research [7]. We have also developed targeted nanosomes against the damaged cartilage by conjugation of nanosomes with a mAb specific to type II collagen (mAbCII) [8]. Since conjugation of antibody to the liposome is a vital factor for proper functioning of the LTLs, therefore, it is of utmost importance that binding of antibody to the liposome is established before its application. Various methods can be utilized to establish this approach such as Enzyme-linked immunosorbent assay (ELISA), Western blot, immunoprecipitation, liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS) and fluorescent microscopy provided the antibody is conjugated to some fluorescent moiety [9]. However, the problem is that these methods are labor intensive and consume time to progress the research work. Therefore, here we present a simple method for the effective quantification of mAb that is bound to liposomes [10]. The liposomes utilized in this study are nano-sized liposomes and hence are referred to as ‘nanosomes’ [8]. Our method provides fast, easy and reliable quantification of bound mAb to nanosomes. The significance of our method lies in the time-effectiveness of the procedure that is a critical factor in every research setting.
2.Materials and Reagents
This procedure described here can be divided in 7 distinct stages: (1) Preparation of Column (Section 3.1); (2) Preparation of nanosomes (Section 3.2); (3) Extrusion of lipid solution (Section 3.3); (4) Size-exclusion chromatography (Section 3.4); (5) Dynamic light scattering (DLS) analysis (Section 3.5); (6) Conjugation of monoclonal antibody and nanosomes (Section 3.6.); (7) Quantification of antibody conjugated to nanosomes (Section 3.7). Protocol for preparation of targeted nanosomes with monoclonal antibody has been adapted from our previously published study [8].
2.1. Materials
- Triton X-100 (BIORAD, Cat#: 1610407)
- 1x Phosphate buffered saline (PBS) (Gibco, Cat#: 10010023)
- Monoclonal anti-type II collagen antibody (mAbCII) (Provided by VA Program Project Scientific Core at the Memphis Veteran Affairs Medical Center, Memphis, TN)
- Normal Mouse IgG (MilliporeSigma, Cat#: 12-371)
- 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) (Avanti Polar Lipids, Inc., Cat#: 850375)
- 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy(polyethylene glycol)-2000] (ammonium salt) (DSPE-PEG2000) (Avanti Polar Lipids, Inc., Cat#: 880120)
- 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[maleimide(polyethylene glycol)-2000] (ammonium salt) (DSPE-PEG2000-Maleimide) (Avanti Polar Lipids, Inc., Cat#: 880126)
- Cholesterol (Avanti Polar Lipids, Inc., Cat#: 700000)
- Chloroform (Fisher Scientific, Cat#: C298-4)
- Sepharose® CL-4B (Sigma, Cat#: CL4B200-100ML)
- Spin Column (Milipore-Sigma, Amicon Ultra, Cat#: UFC510096)
- Ethanol (Sigma, Cat#: E7023)
- Penicillin-Streptomycin (10,000 U/mL) (Gibco, Cat#: 15140122)
- Pierce™ 20X Borate Buffer (Thermo Fisher Scientific, Cat#: 28341)
- Pierce™ Traut's Reagent (2-iminothiolane) (Thermo Fisher Scientific, Cat#: 26101)
- Sodium phosphate monobasic (Sigma, Cat#: S3139)
- Sodium phosphate dibasic (Sigma, Cat#: S3264)
- Distilled water (dH2O)
2.2. Equipment
- Amicon Ultra-0.5 Centrifugal Filter Unit with Ultracel-100 membrane (Millipore, Cat#: UFC510096)
- Eppendorf™ Model 5418 Microcentrifuges (Fisher Scientific, Cat#: 05-403-90)
- Thermo Scientific NanoDrop™ 1000 Spectrophotometer (Thermo Fisher Scientific)
- Nanoparticle Size Analyzer (HORIBA Scientific, Cat#: SZ-100)
- CF-2 Fraction Collector (SPECTRUM, Cat#: 124845)
- Labline 3520 Orbital Shaker (HYLAND SCIENTIFIC)
- DWK Life Sciences Kimble™ Kontes™ FlexColumn™ Economy Columns (Fisher Scientific, Cat#: K420401-1530)
- Fume hood
- Vortex machine
- Nitrogen (N2) Gas Tank with regulator (nexAir, Cat#: UN1066)
- Vacuum chamber
- Pipetman Pipettes (P10, P20, P200, P1000) (GILSON)
- 12 X 75mm Borosilicate Glass Culture Tubes (Baxter, Cat#: T1290-3)
- Sterile transfer pipette (Fisher Scientific, Cat#: 13-711-9CM)
- Mini-Extruder Assembly (Avanti Polar Lipids, Inc.)
- Class A Clear Glass Threaded Vials with Attached Caps, PE Poly-Seal™ Cone Liner (Fisher Scientific, Cat#: 14-955-334)
- PC Membranes 0.2μm (Avanti Polar Lipids, Inc., Cat#: 610006)
- Filter support (Avanti Polar Lipids, Inc., Cat#: 610014)
- Support stand with clamps
- UVette® 220 nm – 1,600 nm, original Eppendorf plastic cuvette (eppendorf, Cat#: 952010051)
- Fisherbrand™ Premium Microcentrifuge Tubes: 1.5mL (Fisher Scientific, Cat#: 05-408-129)
2.3. Software
- NanoDrop 1000 Operating Software, version 3.8.1 (Released June 17, 2011) (Thermo Fisher Scientific)
- HORIBA NextGen Project, SZ-100 for Windows, P2000447001l, 1.90 (Released February 29, 2012) (HORIBA Scientific)
- GraphPad Prism v.5.00 for Windows (GraphPad Software, USA, http://www.graphpad.com)
3.Procedure
3.1. Preparation of the column for size exclusion chromatography
Fix a column (Volume = π x radius2 x length) with a support stand by means of clamps in a straight position over the CF-2 Fraction Collector. Add 5 ml of ethanol to the column, allow it to flow and make sure that there is no leakage and the drops are falling right in the fraction tube. Let the ethanol to run-off completely and turn the knob to the stop position.
Sepharose® CL-4B is already provided as a slurry in 100 ml ethanol. Mix rigorously with hand to completely disperse the Sepharose until a homogenized solution can be observed. Immediately and quickly add 25 ml of Sepharose solution to the column and let it stand overnight at room temperature. This allows a pore size with fractionation range of 30,000-5,000,000 for dextrans and 60,000-20,000,000 for globular proteins.
CRITICAL STEP Make sure that the temperature of the lab remains around the ambient temperature since high temperature can cause bubble formation in the column.
The next day wash the column with freshly prepared PBS supplemented with 2x Penicillin-Streptomycin at least ten times.
CRITICAL STEP PBS with 2x Penicillin-Streptomycin can be prepared by adding 10 ml of Penicillin-Streptomycin (10,000 U/mL) to 490 ml of PBS.
The column is now ready to perform the size-exclusion chromatography.
CRITICAL STEP Keep the column hydrated by adding 5 ml of PBS with 2x Penicillin-Streptomycin.
3.2. Preparation of nanosomes
Prepare the lipid film by mixing DOPC (5.2 μM), cholesterol (4.5 μM), DSPE-PEG2000 (0.3 μM) and DSPE-PEG2000-Maleimide (0.015 μM) at a volume ratio of 52:45:2.9:0.1 in a 10 x 75 mm borosilicate glass culture tube, respectively. The lipid solutions were prepared by dissolving in chloroform/methanol [2:1 (vol/vol)].
CRITICAL STEP Mix well by pipetting and vortex before proceeding to the next step.
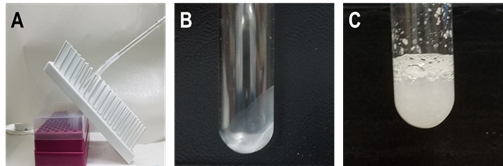
Dry the lipid film by using N2 gas. Keep the tube in tilting position at an angle of 60° until a complete thin dry film is formed (Figure 1A and B).
CRITICAL STEP Make sure to complete dry the lipid film. No liquid should be remaining in the tube before proceeding to the next step.
Place lipid film under vacuum for at least 30 min.
Re-hydrate the lipid film using 400 µl of PBS and mix thoroughly until a homogenized solution is obtained (Figure 1C).
3.3. Extrusion of the lipid solution
Extrusion of the lipid solution will yield nanosomes of appropriate size.
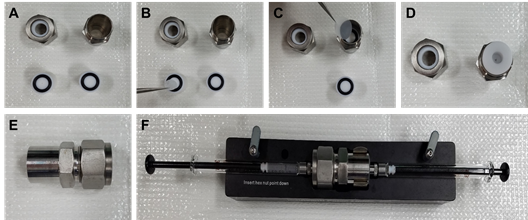
Assemble the extruder assembly as recommended by the manufacturer’s instructions (https://avantilipids.com/divisions/equipment/mini-extruder-assembly-instructions/) (Figure 2). Extrude the lipid film by injecting through the extruder assembly back and forth 21 times.
CRITICAL STEP The size of the membrane used to extrude the lipid mixture depends on the size required by a particular experiment. We used a membrane of 0.2 µm in size to extrude the lipid mixture.
3.4. Size-exclusion chromatography
Size-exclusion chromatography is done to separate the extruded nanosomes from the free molecules.
Let the PBS with 2x Penicillin-Streptomycin that is present in the column for hydration completely flow out before loading the nanosome solution.
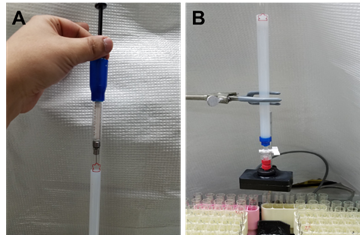
Turn the knob to stop the flow and load the extruded nanosome solution on to the column (Figure 3). Let the nanosome solution to flow into the column.
When no more drops can be observed to come out of the column, add PBS with 2x Penicillin-Streptomycin.
Observe each fraction for the turbidity. The nanosomes of the appropriate size will appear turbid. Select 3-4 fractions that appear to be the most turbid.
Collect all the fractions in a single clear glass vial with a poly-seal cone liner cap (total number of nanosomes per batch is 1.25x1013/ml) calculated by nomogram.
3.5. Dynamic light scattering analysis
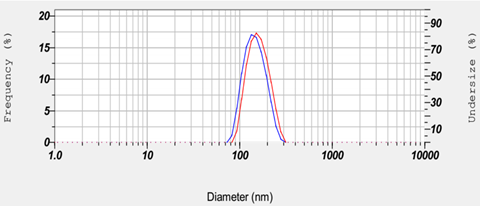
Dynamic light scattering (DLS) is performed to confirm the size of the nanosomes (Figure 4).
Add 10 µl of nanosome solution to 490 µl of PBS in a cuvette and measure the size of the nanosomes using the Nanoparticle Size Analyzer (HORIBA Scientific, Cat#: SZ-100).
Record three values for each sample and take the mean as the average diameter of the nanosomes.
3.6. Conjugation of nanosomes with the monoclonal antibody
Conjugation of the nanosomes can be performed with any desired monoclonal antibody following the method below. In this study, we conjugated the nanosomes with the mAbCII.
Change the buffer of the stock antibody solution to PBS by using a spin column and centrifugation at 12,000 g for 8 min. Collect the stock antibody solution in a new 1.5 ml microcentrifuge tube by inverting the spin column and short spin centrifugation for 5 s.
Thiolate the antibody by suspending the 200 mg of purified monoclonal antibody in 200 μl of 150 mM sodium borate, 0.1 mM EDTA.
PAUSE Note: The final concentration of the antibody is 1 mg/ml.
Mix the mAb and Traut's Reagent (2-iminothiolane) at mAb/Traut’s Reagent [1:20(mol/mol)] and incubate for 1 h at room temperature on an orbital shaker at 1,000 rpm.
CRITICAL STEP Always use fresh stock solution of Traut’s Reagent.
After 1 h, use the spin column to remove excess reactants from the thiolated antibody and change the buffer to 0.1 M sodium phosphate (pH 8.0) by centrifugation at 12,000 g for 8 min.
Mix 150 µl of purified nanosomes with 50 µl of freshly thiolated antibody and incubate the mixture at room temperature overnight with gentle shaking at 1,000 rpm.
Next day perform the size-exclusion chromatography as described previously and also perform DLS analysis.
CRITICAL STEP DLS will slightly differ approximately 10-15 nm in diameter before and after conjugation of the antibody (Figure 4).
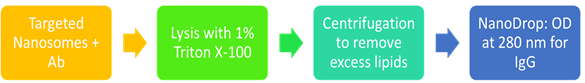
3.7. Quantification of antibody conjugated to the nanosomes
Lyze the targeted nanosomes (sample) with 1% Triton X-100. Briefly, add 1% Triton X-100 to be centrifugation at 12,000 g for 8 min PBS or the buffer (blank) used to prepare the final aliquot of targeted nanosomes and standard antibody solution (standard) of known concentration. Vortex well and leave for 1-2 min at room temperature.
Wash each sample, blank and standard with PBS by centrifugation at 12,000 g for 8 min using a spin column. Repeat this step thrice (3x) to ensure removal of excess lipids and Triton X-100.
CRITICAL STEP Washing is a critical step since presence of lipids and Triton X-100 can result in false positive results.
Collect each sample and standard in a new 1.5 ml microcentrifuge tube by inverting the spin column and short spin centrifugation for 5 s. Dissolve in an appropriate volume of PBS or buffer of choice.
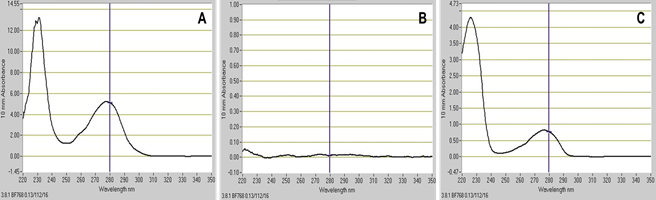
Use the PBS sample as a blank. Measure and record the absorbance at 280 nm for IgG of each sample and standard by using the NanoDrop instrument (Figure 5).
4.Data Analysis
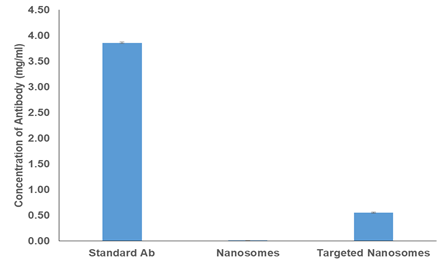
We used the following groups to demonstrate the above procedure: (i) PBS as blank, (ii) antibody standard of known concentration, (iii) nanosomes only to demonstrate that lipids do not interfere with the final readings and (iv) targeted nanosomes to determine the concentration of the bound antibody.
Each sample was treated with Triton X-100, washed with PBS as described in the procedure and dissolved in 100 µl of PBS.
Absorbance was recorded by the NanoDrop instrument at 280 nm for IgG.
Statistical analysis was performed using the GraphPad Prism v.5.00 for Windows (GraphPad Software, USA, http://www.graphpad.com). Data is expressed as mean ± SD, where n = 3 for all the experiments. Student's t test was done to determine statistical significance. Statistical significance was considered at P ≤ 0.05. No background signal can be observed for nanosomes alone (Figure 6 and Figure 7A).
The concentration of standard antibody solution was 4 mg/ml. However, the final mean concentration was 3.86 mg/ml showing that only 3.5% antibody is loss during this procedure. Therefore, the final concentration of the MabCII per nanosome is 0.06 pg/nanosome.
5.Recipes
5.1. Sodium Borate Buffer
Take 7.5 ml of 20X Borate Buffer and make up the final volume up to 50 ml with dH2O to yield 150 mM Sodium Borate buffer.
B. Sodium Phosphate Buffer
0.2 M monobasic stock: 13.9 g sodium phosphate monobasic (NaH2PO4) in 500 mL dH2O.
0.2 M dibasic stock: 28.4 g (Na2HPO4) 1 L dH2O.
0.1 M sodium phosphate buffer: To make 600 ml 0.1 M sodium phosphate buffer (pH 8.0), mix 15.9 ml of 0.2 M monobasic stock with 284.1 of 0.2 M dibasic stock and make up final volume up to 600 ml with dH2O.
Acknowledgements
This work was supported by grants from the Arthritis Foundation (Discovery award, H.C), Oxnard Medical Research Foundation (H.C) and UT Research Foundation (H.C).
Conflicts of Interest: The authors declare no conflict of interest.
Ethics: There are no ethical matters involved in this protocol. This protocol has not contained anything related to human subjects.
References
1. Balazs, D.A. and W. Godbey, Liposomes for use in gene delivery. J Drug Deliv, 2011.:p. 326497.
2. Tiwari, G., et al., Drug delivery systems: An updated review. Int J Pharm Investig, 2012. 2(1): p. 2-11.
3. Voinea, M. and M. Simionescu, Designing of 'intelligent' liposomes for efficient delivery of drugs. J Cell Mol Med, 2002. 6(4): p. 465-74.
4. Puri, A., et al., Lipid-based nanoparticles as pharmaceutical drug carriers: from concepts to clinic. Crit Rev Ther Drug Carrier Syst, 2009. 26(6): p. 523-80.
5. Zamboni, W.C., Liposomal, nanoparticle, and conjugated formulations of anticancer agents. Clin Cancer Res, 2005. 11(23): p. 8230-4.
6. Sercombe, L., et al., Advances and Challenges of Liposome Assisted Drug Delivery. Front Pharmacol, 2015. 6: p. 286.
7. Sapra, P. and T.M. Allen, Ligand-targeted liposomal anticancer drugs. Prog Lipid Res, 2003. 42(5): p. 439-62.
8. Cho, H., et al., Detection of early cartilage damage using targeted nanosomes in a post-traumatic osteoarthritis mouse model. Nanomedicine, 2015. 11(4): p. 939-46.
9. Damen, C.W., J.H. Schellens, and J.H. Beijnen, Bioanalytical methods for the quantification of therapeutic monoclonal antibodies and their application in clinical pharmacokinetic studies. Hum Antibodies, 2009. 18(3): p. 47-73.
10. Bhatti, F., et al., Characterization of Physicochemical and Biological Properties of Type-II Collagen Targeted Nanosomes. Journal of Nanoparticle Research, 2019. 21:235.
11.Liposomes: A Practical Approach (Practical Approach Series (264)), Oxford University Press; 2nd edition (August 7, 2003)
12.Schnyder A, Huwyler J. Drug transport to brain with targeted liposomes. NeuroRx. 2005 Jan;2(1):p. 99-107.
- Bhatti, F U, Heo, S and Cho, H(2021). Quantitation of the Monoclonal Antibody Conjugated to Nanosomes. Bio-protocol Preprint. bio-protocol.org/prep1173.
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link
