Advanced Search
Quantitative real time polymerase chain reaction (qRT-PCR)
Last updated date: Jun 11, 2021 Views: 866 Forks: 0
Quantitative Real-Time Polymerase Chain Reaction assay
Sudhir G. Tattikota1 and Norbert Perrimon1,2
1Department of Genetics, Blavatnik Institute, Harvard Medical School, Boston, MA, USA
2Howard Hughes Medical Institute, Boston MA, USA
Contact: sudhir_gt@hms.harvard.edu
Quantitative real-time PCR is a powerful method used to measure changes in gene expression levels in a particular sample across various conditions or genotypes.
Materials and reagents:
- Primer pairs (typically designed using FlyPrimer Bank (Hu et al., 2013))
- Biological specimen (e.g., blood cells/hemocytes), store at -80°C
- Trizol reagent (ThermoFischer Scientific, catalog number: 15596018), store at 4°C
- Absolute ethanol (Decon Labs, Inc., catalog number: V1016), store at RT
- Nuclease-free water (Ambion, catalog number: AM9930), store at RT
- Schneider’s insect media (Sigma-Aldrich, catalog number: S9895), store at 4°C
- Direct-zol RNA MicroPrep Kit (Zymo Research, catalog number: R2060), store at RT
- Turbo DNA-free Kit (Invitrogen, catalog number: AM1907), store at -20°C
- Pipettes and tips (P2, P20, P200, P1000)
- Eppendorf LoBind tubes (Fischer Scientific, Eppendorf, catalog number: 13-698-794)
- iScript cDNA synthesis Kit (Bio-Rad, catalog number: 1708890), store at-20°C
- Bio-Rad iQ SYBR Green Supermix (Bio-Rad, catalog number: 1708880), store at-20°C
- Bio-Rad Hard-Shell PCR Plates 96-well, thin wall (Bio-Rad, catalog number: HSP9601)
- Bio-Rad Microseal ‘B’ seal Seals (Bio-Rad, catalog number: MSB1001)
Equipment:
- Thermocycler for Polymerase Chain Reaction (PCR)
- Bio-Rad CFX96 Real-Time PCR System (Bio-Rad)
- NanoDrop Microvolume Spectrophotometer (ThermoFischer Scientific)
- Bench top centrifuge
Preparation of hemocytes for RNA isolation:
- Bleed around 50 vortexed larvae (per biological replicate) of desired genotypes directly into 100 ml of Schneider’s media. For preparing larvae for bleeding see (Petraki et al., 2015) and https://bio-protocol.org/prep670
- Transfer the cell suspension into RNase-free Eppendorftubes and place on dry ice (or flash freeze in liquid nitrogen).
- Repeat for other replicates and genotypes and store at -80°C for long term or continue for RNA isolation procedure.
Total RNA isolation:
https://files.zymoresearch.com/protocols/_r2060_r2061_r2062_r2063_direct-zol_rna_microprep.pdf
- Before starting, open the Direct-zol RNA microprep kit and follow the instructions to add absolute ethanol to the RNA wash buffers.
- Carefully label the required number of RNA spin columns with the collection tubes from the kit and keep them ready on a tube rack.
- Retrieve the Eppendorf tubes containing the frozen cell suspension from -80°C and add 300 ml of Trizol to each tube.
- Vortex the tubes and incubate for 5 min at room temperature.
- Add equal volumes of absolute ethanol to the tubes (one tube at a time). Mix well and transfer the entire volume of ~800 ml to the columns that were kept ready in (2).
- Centrifuge the columns at 10,000 g for 30 sec at room temperature.
- Discard the collection tubes and transfer the columns to fresh collection tubes.
- Add 400 ml of RNA pre-wash buffer[Note: Make sure that requiredvolume of ethanolwas added to the buffer prior to use] and spin the columns at 10,000 g for 30 sec at room temperature. Discard the flowthrough.
- Repeat step 8 for one more time.
- Add 700 ml of RNA wash buffer [Note: Make sure that required volume of ethanol was added to the buffer prior to use] and spin the columns at 10,000g for 2 min at room temperature. Discard the flow through and place the columns back in the same collection tubes.
- Centrifuge the columns for an additional 2 min to air dry the RNA spin columns.
- Transfer the columns to RNase free Lobind Eppendorf tubes.
- Add 20 ml of nuclease-free water directly onto the columnsand centrifuge the columns at 10,000 g for 2 min at room temperature.
- Discard the columns and keep the tubes with RNA on ice for downstream DNase treatment.
DNase treatment of total RNA using Turbo DNA-free kit:
https://assets.thermofisher.com/TFS-Assets/LSG/manuals/1907M_turbodnafree_UG.pdf
- Add 2 ml of 10x DNase buffer and 1 ml of DNase enzyme to approximately 17 ml of RNA that is obtained as flowthrough from step 13 of the RNA isolation procedure.
- Mix the samples well and incubate the tubes in a 37°C heat block or water bath for 20 – 30 min.
- Next, add 2 ml of DNase Inactivation Reagent to the samples and incubate for 5 min at room temperature. Make sure to mix the tubes by gently flicking every 2 min.
- Centrifuge the tubes at 10,000 g for 1.5~2 min at room temperature.
- Carefully transfer the supernatant to fresh RNase-free Lobind Eppendorf tubes.
- Quantify the amount of RNA using NanoDrop. Typically, the ratios of both 260/280 nm and 260/230 nm should be close to 1.8~2, which determines high quality RNA.
cDNA synthesis:
https://www.bio-rad.com/webroot/web/pdf/lsr/literature/4106228.pdf
- For the cDNA synthesis, consider using an amount of 500 ng ~ 1 mg of total RNA.
- Transfer the required amount of RNA to nuclease-free PCR compatible strip tubes.
- Make up the volume to 15 ml with nuclease-free water.
- Add 4 ml of 5x iScript reaction mix and 1 ml of iScript reverse transcriptase to each sample.
- Mix well and set up the PCR with the following steps:
- 5 min at 25°C
- 20 min at 46°C
- 1 min at 95°C
- Hold at 4°C (optional)
- Next, add 80 ml of nuclease-free water to the 20 ml of cDNA (1:5 dilution) to prepare a working cDNA stock for downstream qRT-PCR reactions. The cDNA can be stored at 4°C for short duration and at -20°C for long duration of time.
Quantitative Real-Time PCR (qRT-PCR):
- A typical qRT-PCR reaction volume is 10 ml but this volume can also be scaled up to a 15 ml reaction volume set up.
- Thaw the cDNA (if stored at -20°C prior to use), primer pair mix (forward+reverse) and the 2x SYBR Green reagent on ice.
- Prepare master mixes of water, primer pair mix, and SYBR Green to obtain a volume that is sufficient for required number of reactions (including technical replicates for every reaction) with exactly 8 ml total volume per reaction. Below are the volumes required for 1 reaction totaling 8 ml each (Note: Make sure to include 10% of extravolume or 2 extra reactions to account for pipetting error):
- Nuclease free water: 2.95 ml times x-number of reactions
- Primer pair mix (25 mM): 0.05 ml times x-number of reactions
- 2x SYBR Green: 5 ml times x-number of reactions
- Dispense 8 ul of the SYBR Green mix into designated wells of the 96-well Bio-Rad qRT-PCR compatible plate. Repeat for every gene of interest. Include technical replicates for every sample and the gene of interest. For loading the plate, see example below:
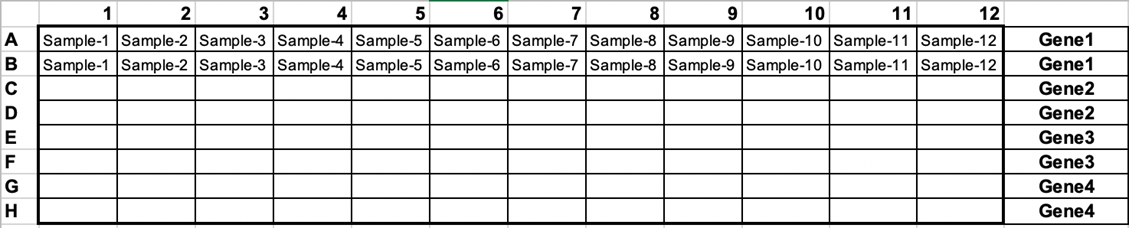
5. Next, add 2 ml of the cDNA working stock to the designated wells (Note: Use a p2 pipette and carefully release the cDNA droplet onto one side of the wall of the PCR plate. Change the tip for every well!).
6. Once the cDNA is added to all designated wells, seal the plate with Bio-Rad Microseal ‘B’ seal.
7. Centrifuge the plate for a quick spin and load the plate onto Bio-Rad CFX96 Real-Time System and start the following program:
- 95°C for 3 min
- 95°C for 10 sec
- 60°C for 30 sec
- Go to step ii for 39 times
- Include melt curve
- End
Data analysis:
For calculating the fold change in gene expression, the Pfaffl’s 2^(-DDCt) method is often used (Pfaffl, 2001). Include multiple housekeeping genes such as Rp49, gapdh, tubulin, etc., to normalize genes of interest.
Useful links:
Preparation of hemocytes in suspension: https://bio-protocol.org/prep670
Primer designing tool for Drosophila genes: https://www.flyrnai.org/flyprimerbank cDNA synthesis: https://www.bio-rad.com/webroot/web/pdf/lsr/literature/4106228.pdf Real-Time PCR experimental design:
https://www.bio-rad.com/en-us/applications-technologies/real-time-pcr-experimental-design?ID=LUSNJVIVK
References:
Hu, Y., Sopko, R., Foos, M., Kelley, C., Flockhart, I., Ammeux, N., Wang, X., Perkins, L., Perrimon, N., and Mohr, S.E. (2013). FlyPrimerBank: an online database for Drosophila melanogaster gene expression analysis and knockdown evaluation of RNAi reagents. G3 Bethesda Md 3, 1607–1616.
Petraki, S., Alexander, B., and Brückner, K. (2015). Assaying Blood Cell Populations of the Drosophila melanogaster Larva. J. Vis. Exp. JoVE.
Pfaffl, M.W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, e45.
- Tattikota, S and Perrimon, N(2021). Quantitative real time polymerase chain reaction (qRT-PCR). Bio-protocol Preprint. bio-protocol.org/prep1155.
- Tattikota, S. G., Cho, B., Liu, Y., Hu, Y., Barrera, V., Steinbaugh, M. J., Yoon, S., Comjean, A., Li, F., Dervis, F., Hung, R., Nam, J., Ho Sui, S., Shim, J. and Perrimon, N.(2020). A single-cell survey of Drosophila blood. eLife. DOI: 10.7554/eLife.54818
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link
