- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Determination of Enzyme Kinetic Parameters of UDP-glycosyltransferases
Published: Vol 3, Iss 14, Jul 20, 2013 DOI: 10.21769/BioProtoc.825 Views: 13088
Reviewed by: Tie Liu

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

A Semi-throughput Procedure for Assaying Plant NADP-malate Dehydrogenase Activity Using a Plate Reader
Kevin Baudry and Emmanuelle Issakidis-Bourguet
Aug 20, 2023 1474 Views

An in vitro Assay to Probe the Formation of Biomolecular Condensates
Yu Zhang and Shen Lisha
Sep 5, 2023 3209 Views

Immunofluorescence for Detection of TOR Kinase Activity In Situ in Photosynthetic Organisms
Ana P. Lando [...] Giselle M. A. Martínez-Noël
Dec 20, 2024 1824 Views
Abstract
The determination of enzyme kinetic parameters, such as the Km and kcat values, is an essential part of the characterization of newly discovered enzymes. This protocol describes the determination of enzyme kinetic parameters of the Barbarea vulgaris UDP-glycosyltransferases (UGTs) UGT73C11 and UGT73C13 toward the sapogenins oleanolic acid and hederagenin as sugar acceptor substrates. UGTs catalyze the transfer of glycosyl residues. They generally use uridine sugar nucleotides as their sugar donor substrates, whereas sugar acceptor substrates arise from structurally diverse sets of metabolite classes. This protocol is based on the quantification of 14C-labeled glycosides following thin layer chromatography (TLC)-based separation. The dependence of the measured signal on a universal radioactively-labeled sugar donor substrate allows the potential application of the protocol in combination with a wide range of different sugar acceptor substrates. However, since the here described TLC separation procedure has been optimized for the separation of sapogenins and their glycosides, some modifications may become necessary when investigating other compound classes.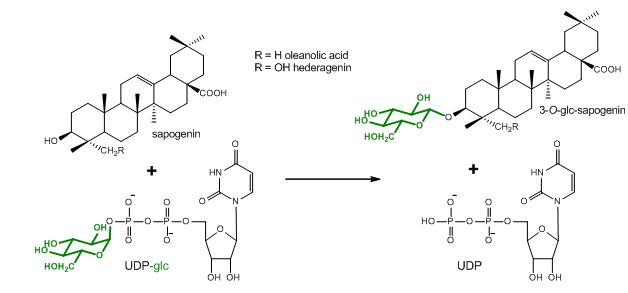
Figure 1. Glucosylation reaction catalyzed by UGT73C10-UGT73C13 from Barbarea vulgaris (Augustin et al., 2012). All four enzymes utilize uridine diphosphate glucose (UDP-glc) as glucosyl-moiety donor substrate and different sapogenins such as the oleanane sapogenins oleanolic acid and hederagenin as glucosyl-moiety acceptor substrates.
Materials and Reagents
- Uridine-5'-diphosphoglucose (UDP-Glc) (e.g. Sigma-Aldrich, catalog number: S451649 )
- Uridine-5'-diphosphate-[14C]glucose (UDP-[14C]Glc) (e.g. PerkinElmer, catalog number: NEC403050UC )
- N-Tris(hydroxymethyl)methyl-3-aminopropanesulfonic acid (TAPS) (e.g. Sigma-Aldrich, catalog number: T5130 )
- Bovine serum albumin (BSA) (e.g. Sigma-Aldrich, catalog number: A7906 )
- Dithiothreitol (DTT) (e.g. Sigma-Aldrich, catalog number: D0632 )
- Silica gel 60 F254 plates (e.g. EMD Millipore, catalog number: 1055540001 )
- Appropriate E. coli protein expression strain (e.g. strain XJb(DE)) (Zymo Research, catalog number: T5051 )
- FRETWorks S-tag assay kit (EMD Millipore, catalog number: 70724 )
- TAPS buffer
- 1.6 mM oleanolic acid stock solution (see Recipes)
- 1.6 mM hederagenin stock solution (see Recipes)
- Pre-master mix for 36 UGT73C11 enzyme assay reactions (see Recipes)
- Pre-master mix for 37 UGT73C13 enzyme assay reactions (see Recipes)
Equipment
- Vacuum centrifuge (e.g. Labogene, catalog number: 7.008.100.777 )
- TLC Developing Chamber (e.g. VWR international, catalog number: 21432-739 )
- Storage phosphor screens (e.g. GE healthcare, catalog number: 28-9564-74 )
- Phosphorimager (e.g. Molecular Dynamics, model: STORM 840 )
Software
- ImageQuant 5.0 (Molecular Dynamics) or similar
- SigmaPlot 11.0 (Systat Software, Inc.) or similar
Procedure
- Preparation of radiolabeled sapogenin-glucoside standards and TLC plates with reference dilution series
- 100 μl (6.623 nmol [74 kBq]) UDP-[14C]Glc were evaporated to dryness in a 1.5 ml microcentrifugation tube using a vacuum centrifuge. The same microcentrifugation tube was used to prepare 200 μl of a glucosylation reaction master mix that was used to generate the reference glucosides.
- Final reaction conditions of the glucosylation reaction were adjusted to 25 mM TAPS pH 8.6, 1 mM DTT, 33.12 μM UDP-[14C]Glc (by dissolving the dried 6.623 nmol) and 467 μM non-radioactive UDP-Glc (additionally added).
- Frozen (-80 °C) E. coli cells resuspended in 10 mM TAPS buffer pH 8.0 were lysed by thawing (see Notes) and insoluble cell debris were removed by centrifugation for 20 min at 20,000 x g, 4 °C.
- The concentration of the recombinant UGTs in the E. coli lysate was determined by applying the FRETWorks S-tag assay kit according to manufacturer's instructions.
- E. coli lysate containing recombinant UGT73C11 was added to the master mix to a final concentration of 200 ng/μl UGT73C11. 60 μl of this master mix were transferred to 0.5 ml microcentrifugation tubes.
- Enzymatic reactions were started by adding 4 μl of a 1.6 mM aglycone stock solution. In our case the applied aglycones were the sapogenins oleanolic acid and hederagenin that had been solubilized in 100% DMSO (thus, final concentrations of the aglycone and DMSO in the enzymatic reaction were 100 μM and 6.25%, respectively).
- Complete conversion of the supplied aglycones to their corresponding glucosides was achieved by incubating the reactions overnight (18 h) at 37 °C (shaking is not necessary for this step).
- Conversion efficiency was evaluated by TLC analysis. For this purpose both sapogenin-glucosides and possibly remaining sapogenins were 4 times extracted with 50 μl ethyl acetate from a 20 μl aliquot of the complete reaction.
- The merged ethyl acetate fractions were evaporated to dryness in a vacuum centrifuge.
- The dried extracts were dissolved in 20 μl 96% ethanol and this solution stepwise (3.5 μl per step) spotted to a silica gel TLC plate. The TLC plate was pre-run for 1-2 min using methanol as mobile phase until the solvent front was approximately 1 cm above the loading line.
- The methanol was left to evaporate in a fume hood and the dry plate subsequently developed using dichloromethane: methanol: water (80:19:1) as mobile phase. After evaporation of the mobile phase the developed plate was sprayed with 10% sulfuric acid in methanol and heated to 100 °C until sapogenin and sapogenin-glucosides became visible as reddish bands on the TLC plate.
- The stained TLC plate was evaluated under visible as well as under long wave UV (366 nm) light.
- To generate standard curves of the 14C-labled sapogenin-glucosides, sequential dilutions of the glucosidation reactions were made.
- The highest chosen sapogenin-glucoside concentration for the standard curve was a 1:5 dilution of the overnight reaction with 62.5% ethanol, corresponding to a 20 μM sapogenin-glucoside concentration.
- This dilution was 11 times sequentially diluted with 50% ethanol, thereby generating dilutions with sapogenin-glucoside concentrations of 10 μM, 5 μM, 2.5 μM, 1.25 μM, 0.625 μM, 0.313 μM, 0.156 μM, 0.078 μM, 0.039 μM, 0.020 μM and 0.010 μM.
- 20 μl of each dilution were in 5 μl steps loaded to a silica gel TLC plate, which corresponds to sapogenin-glucoside amounts per spot of 400 nmol, 200 nmol, 100 nmol, 50 nmol, 25 nmol, 12.5 nmol, 6.25 nmol, 3.13 nmol, 1.56 nmol, 0.78 nmol, 0.39 nmol and 0.20 nmol, respectively.
- After evaporation of all solvents the TLC plates were pre-run for 1-2 min using methanol as mobile phase until the solvent front was approximately 1 cm above the loading line.
- The methanol was left to evaporate in a fume hood, and the TLC plates were afterwards developed with dichloromethane: methanol: water (80:19:1) as mobile phase.
- All solvents were left to evaporate and 1.5 μl, 1.0 μl and 0.5 μl of the original master mix of the glucosylation reaction (before addition of the sapogenins) were spotted on an unused side lane of the TLC plate. These spots would allow at a later point normalization of variation in the radioactivity between different reaction master mixes.
Notes:- Local regulations may require you to work with radioactive compounds in specialized areas. Please inform yourself about regulations and guidelines of working with radioactive compounds that apply at your workplace.
- UDP-[14C]Glc was dried out prior to usage, since Perkin Elmer provides this reagent solubilized in 70% ethanol, which serves as a sugar acceptor substrate for the investigated UGTs itself.
- Recombinant UGTs were in this case expressed in the BL21(DE) derivative XJb(DE). This strain expresses a viral endolysin protein and thus allows to be lysed by simply thawing after being frozen.
- Quantification with the FRETWorks S-tag assay kit is based on regeneration of RNaseS activity due the interaction of the S protein (included in the kit) and the S-tag N-terminally fused to the recombinant expressed UGTs. See kit manual for additional information.
- As references for the unmodified sapogenins were 2 nmol oleanolic acid and hederagenin loaded to the TLC plate that was used to determine the conversion efficiency.
- Sapogenin/saponin-glucoside evaluation on sulfuric acid stained TLC plates under long wave UV light is approximately 10 times more sensitive than under visible light.
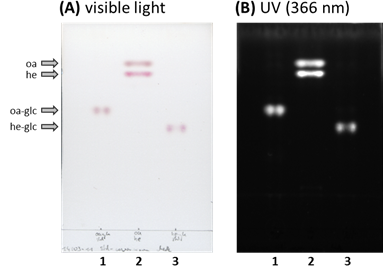
Figure 2. TLC plate with aliquots of the reactions to generate radiolabled (1) 3-O-[14C]glc-oleanolic acid (oa-glc) or (3) 3-O-[14C]glc-hederagenin (he-glc) standards. The TLC plate was evaluated under (A) visible (colored) as well as under (B) long wave UV light (366 nm, black/white). For comparison purpose were authentic (oa) oleanolic acid and (he) hederagenin (2 nmol each) loaded to lane (2). - Local regulations may require you to work with radioactive compounds in specialized areas. Please inform yourself about regulations and guidelines of working with radioactive compounds that apply at your workplace.
- 100 μl (6.623 nmol [74 kBq]) UDP-[14C]Glc were evaporated to dryness in a 1.5 ml microcentrifugation tube using a vacuum centrifuge. The same microcentrifugation tube was used to prepare 200 μl of a glucosylation reaction master mix that was used to generate the reference glucosides.
- Preparation of enzyme assays for the determination of enzyme kinetic parameters
- In preparation of the UGT73C11 enzyme assays were 228 μl UDP-[14C]Glc (15.10 nmol [168.72 kBq]) evaporated to dryness in a 1.5 ml microcentrifugation tube using a vacuum centrifuge.
- 2.39 μl and 2.18 μl of a 1 mM UDP-Glc (non-radioactive) solution were evaporated to dryness in two additional 1.5 ml microcentrifugation tubes. Simultaneously, were for the UGT73C13 assays 86.4 μl UDP-[14C]Glc (5.72 nmol [63.94 kBq]), 1.99 μl 0.5 mM UDP-Glc (non-radioactive) and 1.59 μl 0.25 mM UDP-Glc (non-radioactive) dried out in 1.5 ml microcentrifugation tubes.
- Of the acceptor substrates, oleanolic acid and hederagenin, were for the UGT73C11 assays stock solutions with concentrations of 128 μM, 96 μM, 64 μM, 32 μM, 16 μM, 8 μM, 4 μM and 2 μM in 100% DMSO prepared.
- For the UGT73C13 assays were acceptor substrate stock solutions of 1,600 μM, 1,200 μM, 800 μM, 400 μM, 200 μM, 100 μM, 50 μM and 25 μM prepared.
- Frozen (-80 °C) aliquots of E. coli cells resuspended in 10 mM TAPS buffer pH 8.0 were lysed by thawing (see Notes) and insoluble cell debris removed by centrifugation for 20 min at 20,000 x g, 4 °C.
- The concentration of the recombinant UGTs in the E. coli lysate was determined by applying the FRETWorks S-tag assay kit according to manufacturer's instructions.
- The E. coli lysates were diluted to either 5 ng/μl (UGT73C11) or 50 ng/μl (UGT73C13) recombinant protein with 10 mg/ml BSA in 10 mM TAPS buffer pH 8.0.
- In preparation of the UGT73C11 enzyme assays were 228 μl UDP-[14C]Glc (15.10 nmol [168.72 kBq]) evaporated to dryness in a 1.5 ml microcentrifugation tube using a vacuum centrifuge.
- Enzyme assays for UGT73C11
- Preparation of pre-master mix for 36 enzyme assay reactions as described in the recipe section.
- 427.4 μl of this pre-master mix were added to the microcentrifuge tube that contained the dried UDP-[14C]Glc to prepare a reaction master mix that would result with concentrations of the sugar donor substrate of 33.12 μM UDP-[14C]Glc (0.37 kBq/μl) and 467 μM non-radioactive UDP-Glc in the final enzyme assay.
- 96 μl of this master mix were transferred to the tube in which 2.39 μl 1 mM UDP-Glc had been dried out and mixed with 144 μl of the original pre-master mix. Enzyme assays set up with this master mix would have a sugar donor substrate concentration of 13.25 μM UDP-[14C]Glc (0.15 kBq/μl) and 487 μM non-radioactive UDP-Glc.
- 82 μl of the latter master mix were transferred to the tube in which 2.18 μl 1 mM UDP-Glc had been dried out, and mixed with 82 μl of the original pre-master mix. This last master mix is for preparing enzyme assays with a concentration of 6.62 μM UDP-[14C]Glc (0.08 kBq/μl) and 493 μM non-radioactive UDP-Glc.
- 18.75 μl aliquots of the master mixes were transferred to 1.5 ml microcentrifuge tubes on ice.
- For performing the actual assays, tubes with master mix aliquots were pre-incubated for 3 min at 30 °C and the enzymatic reaction started by addition of 1.25 μl acceptor substrate stock solutions prepared in step B-3.
- The master mix with the highest concentration of UDP-[14C]Glc (33.12 μM) was used to assay UGT73C11 with final sugar acceptor substrate concentrations of 1 μM, 0.5 μM, 0.25 μM and 0.125 μM, by adding 1.25 μl of the 16 μM, 8 μM, 4 μM and 2 μM oleanolic acid or hederagenin stock solutions.
- Similarly was the 13.25 μM UDP-[14C]Glc master mix used for final concentrations of 4 and 2 μM oleanolic acid and hederagenin, and the 6.62 μM UDP-[14C]Glc master mix for final concentrations of 8 and 6 μM of the two acceptor substrates. The corresponding stock solutions in these cases were 64 μM, 32 μM, 128 μM and 96 μM.
- The master mix with the highest concentration of UDP-[14C]Glc (33.12 μM) was used to assay UGT73C11 with final sugar acceptor substrate concentrations of 1 μM, 0.5 μM, 0.25 μM and 0.125 μM, by adding 1.25 μl of the 16 μM, 8 μM, 4 μM and 2 μM oleanolic acid or hederagenin stock solutions.
- After addition of the acceptor substrate, enzymatic reactions were allowed to take place by incubation for 3 min at 30 °C, and subsequently stopped by addition of 50 μl ethyl acetate and vigorous mixing for 10 sec.
- Preparation of pre-master mix for 36 enzyme assay reactions as described in the recipe section.
- Enzyme assays for UGT73C13 (in general similar to procedure C, however, enzyme and substrate concentrations differ, as UGT73C13 is less efficient in catalyzing the investigated reaction)
- Preparation of pre-master mix for 37 enzyme assay reactions as described in the recipe section.
- 405 μl of the pre-master mix were transferred to the tube with the dried UDP-[14C]Glc to prepare a reaction master mix for enzyme assays with a final concentration of 13.25 μM UDP-[14C]Glc (0.15 kBq/μl) and 487 μM non-radioactive UDP-Glc.
- 150 μl of this master mix were mixed with 150 μl of the pre-master mix in the microcentrifugation tube in which 1.99 μl 0.5 mM UDP-Glc had been dried out. This master mix was applied for reactions with a final sugar donor substrate concentrations of 6.62 μM UDP-[14C]Glc (0.08 kBq/μl) and 493 μM non-radioactive UDP-Glc.
- For a master mix for enzyme assays with a final UDP-[14C]Glc concentration of 3.31 μM (0.04 kBq/μl) and 497 μM non-radioactive UDP-Glc 120 μl of the previous master mix were diluted with 120 μl of the pre-master mix in the tube that contained the dried out 1.59 μl 0.25 mM UDP-Glc. The 13.25 μM UDP-[14C]Glc master mix was applied for assays with final oleanolic acid and hederagenin concentrations of 1.56 μM, 3.12 μM and 6.25 μM.
- 1.25 μl of the 25 μM, 50 μM and 100 μM sugar acceptor substrate stock solutions were added to 18.75 μl master mix to adjust these sugar acceptor substrate concentrations. Similarly, was the 6.62 μM UDP-[14C]Glc master mix used to assay final sugar acceptor substrate concentration of 12.5 μM and 25 μM (200 μM and 400 μM stock solutions), and the 3.31 μM UDP-[14C]Glc master mix for sugar acceptor substrate concentrations of 50 μM, 75 μM and 100 μM (800 μM, 1,200 μM and 1,600 μM stock solutions).
- Preparation of pre-master mix for 37 enzyme assay reactions as described in the recipe section.
- Analysis of the enzyme assays
- Stopped assays were 4 times extracted with 50 μl ethyl acetate and the merged ethyl acetate extracts evaporated to dryness in a vacuum centrifuge.
- The dried extracts were dissolved in 20 μl 96% ethanol and in 5 μl steps loaded to silica gel TLC plates.
- Potential remainders of the radioactive sapogenin-glucosides were washed out with another 20 μl 96% ethanol and loaded to the same spots on the TLC plates.
- The TLC plates were pre-run for 1-2 min using methanol as mobile phase until the solvent front was approximately 1 cm above the loading line.
- The methanol was left for evaporation in a fume hood and the dried TLC plates were afterwards developed using dichloromethane: methanol: water (80:19:1) as mobile phase.
- After evaporation of the mobile phase, 0.5 μl, 1.0 μl and 1.5 μl of the master mix with the highest UDP-[14C]Glc were spotted on an unused lane of the TLC plate. Additionally, were also 1 μl of the two remaining master mixes with the lower UDP-[14C]Glc spotted to each TLC plate.
- The dried TLC plates were together with one of the prepared reference dilution series TLC plates of the same sapogenin-glucoside exposed for several days to a storage phosphor screen.
- The exposed phosphor screens were scanned with a STORM 840 Phosphorimager and signal intensities in the digital scans quantified using ImageQuant 5.0.
- To compensate for the 10-fold lower concentration of the radioactive sugar donor substrate as compared to the master mix used for generating the reference glucosides, were all sapogenin-glucoside intensities resulting from assays with a 3.31 μM UDP-[14C]Glc master mix prior to their further evaluation multiplied by factor 10. Similarly, were sapogenin-glucoside intensities resulting from 13.25 μM and 6.62 μM UDP-[14C]Glc master mixes multiplied by 2.5 and 5, respectively.
- To further address unintended variation in the amounts of UDP-[14C]Glc in the master mixes used for generating the reference glucosides and the actual enzyme assays, all glucoside intensities of the assays were multiplied with the ratio between the intensities of the corresponding master mix spots on each TLC plate.
- Km and Vmax values were calculated using SigmaPlot 11.0 to perform non-linear regression according to the Michaelis-Menten equation or the velocity equation for substrate inhibition (please refer to the SigmaPlot manual for an instruction how to perform regressions).
Notes:- Recombinant UGTs were in this case expressed in the BL21(DE) derivative XJb(DE). This strain expresses a viral endolysin protein and thus allows to be lysed by simply thawing after being frozen.
- Upon lysis of the E. coli cells all lysates, lysate dilutions and master mixes were constantly kept on ice.
- Quantification with the FRETWorks S-tag assay kit is based on regeneration of RNaseS activity due the interaction of the S protein (included in the kit) and the S-tag N-terminally fused to the recombinant expressed UGTs. See kit manual for additional information.
- The E. coli lysates were diluted with a BSA solution instead of pure buffer, since the specific activity of the recombinant UGTs was seen to decrease upon reduction of total protein concentration.
- The final amount of the investigated enzyme should be chosen based on its activity. If too much of the acceptor substrate is used up within the applied incubation time, the determined reaction velocity will not be in the initial linear range anymore.
- The intention behind using master mixes with different ratios of radiolabeled to non-labeled UDP-Glc was to use as low amounts of the radiolabeled UDP-[14C]Glc as possible, while still ensuring sufficient signal strength for assays with the lowest acceptor substrate concentrations. The final total concentration of 500 μM UDP-Glc was kept throughout the experiment to provide saturating conditions of the sugar donor substrate.
- The different pH in assays of UGT73C11 and UGT73C13 were chosen due to different pH optima of the two enzymes.
- Which concentrations of the acceptor substrate are tested, depends on the investigated enzyme. For deciding optimal concentrations, it is helpful to estimate the Km value in pre-experiments and choose concentrations below and above the estimated Km value for the final experiment.
- The optimal reaction time is dependent on the enzyme activity and the enzyme amount. Time course experiments have to be performed to determine if the reaction velocity is still in the initial linear range after the chosen incubation time. If too much of the acceptor substrate is converted at the lowest applied concentrations, resulting v/S-characteristics are typically sigmoid instead of hyperbolic.
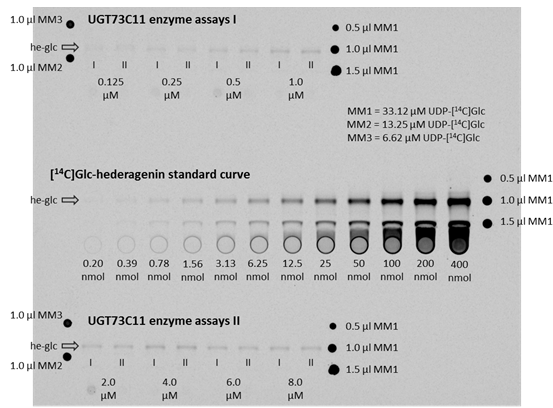
Figure 3. Autoradiogram of TLC plates to determine kinetic parameters of UGT73C11 towards hederagenin. The TLC plates with the ethyl acetate extracts from the actual enzyme assays are on the top as well as on the bottom, whereas the co-exposed TLC plate with the [14C]Glc-hederagenin standard curve is located in the middle. The band representing 3-O-[14C]Glc-hederagenin is for each plate marked with an arrow and the label he-glc. The concentrations below the he-glc bands of the UGT73C11 enzyme assay extracts represent the hederagenin concentration in the corresponding enzyme assay, while I and II mark duplicates. Substance amounts below each standard curve lane represent the amount of [14C]Glc-hederagenin present in the corresponding lane. Master mix aliquots to account for variation in the amount of UDP-[14C]Glc in the master mix preparation (MM1, MM2, MM3) were spotted either right or left of lanes with the actual assays or standards. - Recombinant UGTs were in this case expressed in the BL21(DE) derivative XJb(DE). This strain expresses a viral endolysin protein and thus allows to be lysed by simply thawing after being frozen.
- Stopped assays were 4 times extracted with 50 μl ethyl acetate and the merged ethyl acetate extracts evaporated to dryness in a vacuum centrifuge.
Recipes
- 1.6 mM oleanolic acid stock solution
Dissolve 7.30 mg oleanolic acid in 10 ml DMSO - 1.6 mM hederagenin stock solution
Dissolve 7.56 mg hederagenin in 10 ml DMSO - Pre-master mix for 36 UGT73C11 enzyme assay reactions
36 μl 500 mM TAPS buffer pH 8.6
3.6 μl 200 mM DTT
36 μl 9.34 mM UDP-Glc
72 μl 10 mg/ml BSA
72 μl 5 ng/μl UGT73C11 (diluted E. coli lysate)
455.4 μl water - Pre-master mix for 37 UGT73C13 enzyme assay reactions
37 μl 500 mM TAPS buffer pH 7.9
37 μl 200 mM DTT
37 μl 9.34 mM UDP-Glc
74 μl 10 mg/ml BSA
74 μl 50 ng/μl UGT73C13 (diluted E. coli lysate)
465.05 μl water
Acknowledgments
This protocol was adapted and modified from various, previous 14C-UDP-glucose-based UGT enzyme assay protocols commonly applied in the Section for Plant Biochemistry – Department for Plant Biochemistry Biotechnology – Faculty of Life Sciences – University of Copenhagen and preceding organizations. This work was supported by the Danish Council for Independent Research, Technology, and Production Sciences (grant nos. 09–065899/FTP and 274–06–0370), by the Villum Kann Rasmussen Foundation to Pro-Active Plants, and by a PhD stipend from the Faculty of Life Sciences, University of Copenhagen (to J.M.A.).
References
- Augustin, J. M., Drok, S., Shinoda, T., Sanmiya, K., Nielsen, J. K., Khakimov, B., Olsen, C. E., Hansen, E. H., Kuzina, V., Ekstrom, C. T., Hauser, T. and Bak, S. (2012). UDP-glycosyltransferases from the UGT73C subfamily in Barbarea vulgaris catalyze sapogenin 3-O-glucosylation in saponin-mediated insect resistance. Plant Physiol 160(4): 1881-1895.
Article Information
Copyright
© 2013 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Augustin, J. M. and Bak, S. (2013). Determination of Enzyme Kinetic Parameters of UDP-glycosyltransferases. Bio-protocol 3(14): e825. DOI: 10.21769/BioProtoc.825.
- Augustin, J. M., Drok, S., Shinoda, T., Sanmiya, K., Nielsen, J. K., Khakimov, B., Olsen, C. E., Hansen, E. H., Kuzina, V., Ekstrom, C. T., Hauser, T. and Bak, S. (2012). UDP-glycosyltransferases from the UGT73C subfamily in Barbarea vulgaris catalyze sapogenin 3-O-glucosylation in saponin-mediated insect resistance. Plant Physiol 160(4): 1881-1895.
Category
Plant Science > Plant biochemistry > Protein > Activity
Biochemistry > Protein > Activity
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










