- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
RNA-Seq Library Generation from Rare Human Cells Isolated by FACS
Published: Vol 3, Iss 12, Jun 20, 2013 DOI: 10.21769/BioProtoc.791 Views: 13131
Reviewed by: Lin Fang

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

snPATHO-seq: A Detailed Protocol for Single Nucleus RNA Sequencing From FFPE
Wani Arjumand [...] Luciano G. Martelotto
May 5, 2025 1961 Views

RACE-Nano-Seq: Profiling Transcriptome Diversity of a Genomic Locus
Lu Tang [...] Philipp Kapranov
Jul 5, 2025 2540 Views

Analyzing RNA Localization Using the RNA Proximity Labeling Method OINC-seq
Megan C. Pockalny [...] J. Matthew Taliaferro
Aug 5, 2025 2374 Views
Abstract
High throughput RNA Sequencing has revolutionized transcriptome analyses. However, most available protocols require micrograms of RNA rendering this technique not feasible for analyzing small numbers of cells, including precious rare cell types isolated from human tissues or organs. Here, we used an RNA Amplification System and describe a method for preparing RNA sense-strand cDNA libraries compatible with an Illumina sequencing platform starting from limited numbers of human fetal germ cells as well as human embryonic stem cells (hESCs) isolated using Fluorescence Activated Cell Sorting (FACS). With this protocol we generated seven RNA-Seq libraries starting from 4,000 germ cells sorted from fetal ovaries (n = 2) and fetal testes (n = 2) at 16-16.5 weeks of development and 4,000 sorted hESCs (n = 3). We predict that multiplexed libraries can also be generated by replacing the single-plex 3’ adapter used here with a multiplexing compatible 3’ adapter and indexed PCR primers.
Keywords: PGCMaterials and Reagents
- RNeasy Micro Kit (QIAGEN, catalog number: 74004 )
- WT-Ovation Pico RNA Amplification System (Nugen, catalog number: 3300 )
- WT-Ovation Exon Module (Nugen, catalog number: 2000 )
- Qubit dsDNA HS Assay Kit (Life Technologies, Invitrogen™, catalog number: Q32851 )
- Tris-EDTA pH 8.0, molecular biology grade (Life Technologies, Ambion®, catalog number: AM9849 )
- TruSeq DNA Sample Preparation Kit (Illumina, catalog number: FC-121-2001 )
- MinElute PCR Purification Kit (QIAGEN, catalog number: 28004 )
- QIAquick Gel Extraction Kit (QIAGEN, catalog number: 28704 )
- Certified Low Range Ultra Agarose (Bio-Rad Laboratories, catalog number: 161-3107 )
Equipment
- BD FACSAria Fluorescence activated cell sorter
- NanoDrop 1000
- PCR Thermal Cycler
- Sonic Dismembrator 550 (Thermo Fisher Scientificc)
- Microtip (Misonix Inc, catalog number: 419 )
- Fluorometer
- Agarose gel running apparatus and UV-light transilluminator
- Illumina HiSeq 2000

Figure 1. Overview of the protocol and main reagents used
Procedure
- Sort 4,000 cells directly in 75 μl of RLT buffer (Qiagen RNeasy Micro Kit) using BD FACSAria cell sorter.
- Isolate total RNA from sorted cells using the RNeasy Micro Kit according to manufacturer’s protocol without the addition of β-mercaptoethanol to RLT buffer. Perform on column DNA digestion for 15 min according the manufacturer’s instructions using the DNase that is provided with the RNeasy Micro Kit and elute in 14 μl of RNase/DNase free water. Yield of total RNA using a NanoDrop 1000 ranges between 30-80 ng.
- Amplify RNA and generate cDNA with complementary sequences to the original mRNA using the WT-Ovation Pico RNA Amplification System according to manufacturer’s instruction. Expected yield of amplified cDNA measured on a NanoDrop 1000 is between 5-10 μg. A quality control step can be included at this point by performing Real-Time PCR for known transcripts that are expected to be present in the amplified cDNA pool.
- Generate sense-strand cDNA targets from 3 μg of complementary sequence cDNA using the WT-Ovation Exon Module kit according to manufacturer’s instructions.
- Sonicate 3 μg of sense-strand cDNA that is diluted to a final volume of 400 μl Tris-EDTA pH 8.0 buffer to generate cDNA fragments in the 200-500 bp range. Sonication with Sonic Dismembrator 550 and a 419 Microtip is performed on ice with six pulses of 20 sec each (magnitude setting of 3) and a 60 sec rest interval.
- Quantify sense-strand sonicated cDNA on a fluorometer using the Qubit HS dsRNA assay kit according to manufacturers’ instructions and aliquot 30 ng for generating the Sequencing library.
- Perform End Repair (Reagents are part of the TruSeq DNA Sample Preparation Kit)
20 °C 30 min on a thermal cyclerSense-strand cDNA (30 ng) 30 μl Water 10 μl T4 DNA Ligase Buffer with 10 mM ATP 5 μl 10 mM dNTP mix 2 μl T4 DNA Polymerase (3 U μl-1) 1 μl Klenow DNA Polymerase (1 U μl-1 diluted with water) 1 μl T4 PNK (10 U μl-1) 1 μl - Purify using the MinElute PCR Purification Kit according to manufacturer’s instructions and elute in 34 μl of RNase/DNase free water. Use all eluted cDNA in following step.
- Adenylate 3’ ends (Reagents are part of the TruSeq DNA Sample Preparation Kit)37 °C 30 min on a thermal cycler
cDNA 34 μl 10x Klenow buffer 5 μl 1 mM dATP 10 μl Exo-Klenow (5 U μl-1) 1 μl - Purify using the MinElute PCR Purification Kit according to manufacturer’s instructions. Elute with 10 μl of RNase/DNase free water.
- Ligate paired-end adapters (reagents are part of the TruSeq DNA Sample Preparation Kit)Room temperature 15 min
cDNA 10 μl 2x Ligase buffer 15 μl Adapter oligo mix (1/10 dilution in water) 1 μl DNA ligase 4 μl - Purify the ligation reaction using the MinElute PCR Purification Kit according to manufacturer’s instructions.
- Size select the cDNA library by running the purified PCR product through a 1.5% Agarose Gel, cutting a smear in the range of 150-300 bp, purifying using the QIAquick Gel Extraction Kit according to manufacturer’s instructions and eluting with 36 μl of RNase/DNase free water. Note that DNA concentration is very low therefore there is no visible DNA band on the gel.
- PCR amplification of the adapter modified DNA fragments (Reagents are part of the TruSeq DNA Sample Preparation Kit)Use the following protocol on a thermal cycler
DNA 36 μl 5x Phusion buffer 10 μl 10 mM dNTPs 1.5 μl PCR primer 1.1 1 μl PCR primer 1.2 1 μl Phusion DNA Polymerase (2 U μl-1) 0.5 μl - 98 °C 30 sec
- 18 cycles of
98 °C 10 sec
65 °C 30 sec
72 °C 30 sec - 72 °C 5 min
- 4 °C hold
- 98 °C 30 sec
- Validate library by running the amplified DNA through a 1.5% Agarose Gel. The majority of the amplified cDNA should appear between the 150-300 bp range. Primer dimers and adapters appear at the 30-60 bp range.
- Purify library by gel extracting the DNA that appears in the 150-300 bp range using the QIAquick Gel Extraction Kit according to manufacturer’s instructions and eluting with 15 μl RNase/DNase free water (Figure 2). Quantify library final yield on a fluorometer using the Qubit HS dsDNA assay according to manufacturer’s instructions. The libraries generated using this protocol yielded between 200-700 ng.
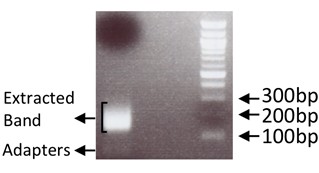
Figure 2. Example of gel extracted band alongside 100 bp ladder. Adapters visible around 30-60 bp. - A quality control step can be included at this point by performing Real-Time PCR for known transcripts that are expected to be present in the generated library. If a quality control step was performed at step 3, before generating the library, the results should be similar.
- Run the cDNA library on an Illunina HiSeq 2000. Alignment statistics for all samples analyzed as well as correlation scores on the 3 replicates of hESC RNA-Seq libraries are shown in Tables 1 and 2.
Table 1. Alignment statisticsSample Total reads Uniquely aligned reads (% total) Testis 16 wk 116,399,186 35,497,900 30.5 Testis 16.5 wk 153,982,474 37,907,565 24.6 Ovary 16 wk 102,837,931 44,784,216 43.5 Ovary 16.5 wk 211,659,065 93,766,963 44.3 hESCs 1 161,037,803 39,339,764 24.4 hESCs 2 133,355,463 38,571,046 28.9 hESCs 3 104,997,235 37,164,448 35.4
Table 2. Correlation scores for hESC RNA-Seq librarieshESCs 1 hESCs 2 hESCs 3 hESCs 1 1 0.96 0.96 hESCs 2 0.97 1 0.97 hESCs 3 0.96 0.97 1
Acknowledgments
A.T.C. is supported by funds from the NIH/NICHD 2 R01 HD058047.
References
- Gkountela, S., Li, Z., Vincent, J. J., Zhang, K. X., Chen, A., Pellegrini, M. and Clark, A. T. (2013). The ontogeny of cKIT+ human primordial germ cells proves to be a resource for human germ line reprogramming, imprint erasure and in vitro differentiation. Nat Cell Biol 15(1): 113-122.
Article Information
Copyright
© 2013 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Gkountela, S. and Clark, A. T. (2013). RNA-Seq Library Generation from Rare Human Cells Isolated by FACS. Bio-protocol 3(12): e791. DOI: 10.21769/BioProtoc.791.
Category
Systems Biology > Transcriptomics > RNA-seq
Molecular Biology > RNA > RNA sequencing
Stem Cell > Embryonic stem cell
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link