- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
On-Column Dual-Gradient Refolding for Efficient Recovery of Insoluble Affinity-Tagged Recombinant Proteins
Published: Vol 16, Iss 3, Feb 5, 2026 DOI: 10.21769/BioProtoc.5598 Views: 93
Reviewed by: Anonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

A Novel Protein Purification Approach Using Elastin-Like Polypeptides (ELP) With His-Tag Assistance
Young Kee Chae and Han Bin Shin
Jun 20, 2025 3128 Views

Thermus thermophilus CRISPR Cas6 Heterologous Expression and Purification
Junwei Wei [...] Yingjun Li
Jul 20, 2025 2172 Views

Prokaryotic Expression and Purification of the hSox2-HMG Domain
Lijie Yang [...] Jingjun Hong
Aug 20, 2025 2386 Views
Abstract
This article presents an efficient protocol for refolding recombinant proteins that are prone to aggregation and form inclusion bodies during expression in Escherichia coli. As a model system, the homolog of CRISPR-associated effector protein CasV-M was investigated. The key element of the developed approach is refolding directly on a metal-affinity Ni-TED (N,N,N´-tris(carboxymethyl)ethylendiamine) resin using a dual-gradient system: a stepwise reduction in the concentration of the chaotropic agent combined with a simultaneous increase in the concentration of a mild nonionic detergent. This combination ensures spatial separation of protein molecules, minimizes aggregation, and promotes the recovery of the native conformation. The resulting method appears to be an alternative to conventional refolding strategies, with potential improvements in the reproducibility and yield of soluble protein compared to dialysis or dilution. The proposed approach can be extended to a broad range of aggregation-prone proteins and is considered a promising strategy for obtaining otherwise insoluble recombinant proteins.
Key features
• This protocol requires optimizing E. coli protein expression and an FPLC system, being particularly suitable for insoluble proteins that form inclusion bodies.
• The method utilizes a dual-gradient refolding strategy on a Ni-TED column, integrating solubilization, refolding, and initial purification into a single workflow.
• Effectively rescues challenging proteins from inclusion bodies, demonstrated with aggregation-prone CRISPR-associated effector homologs (Cas12m) refractory to traditional refolding methods.
• Utilizes Ni-TED resin for its compatibility with high β-mercaptoethanol concentrations and moderate binding affinity, reducing nonspecific binding of host cell proteins.
Keywords: Recombinant proteinGraphical overview
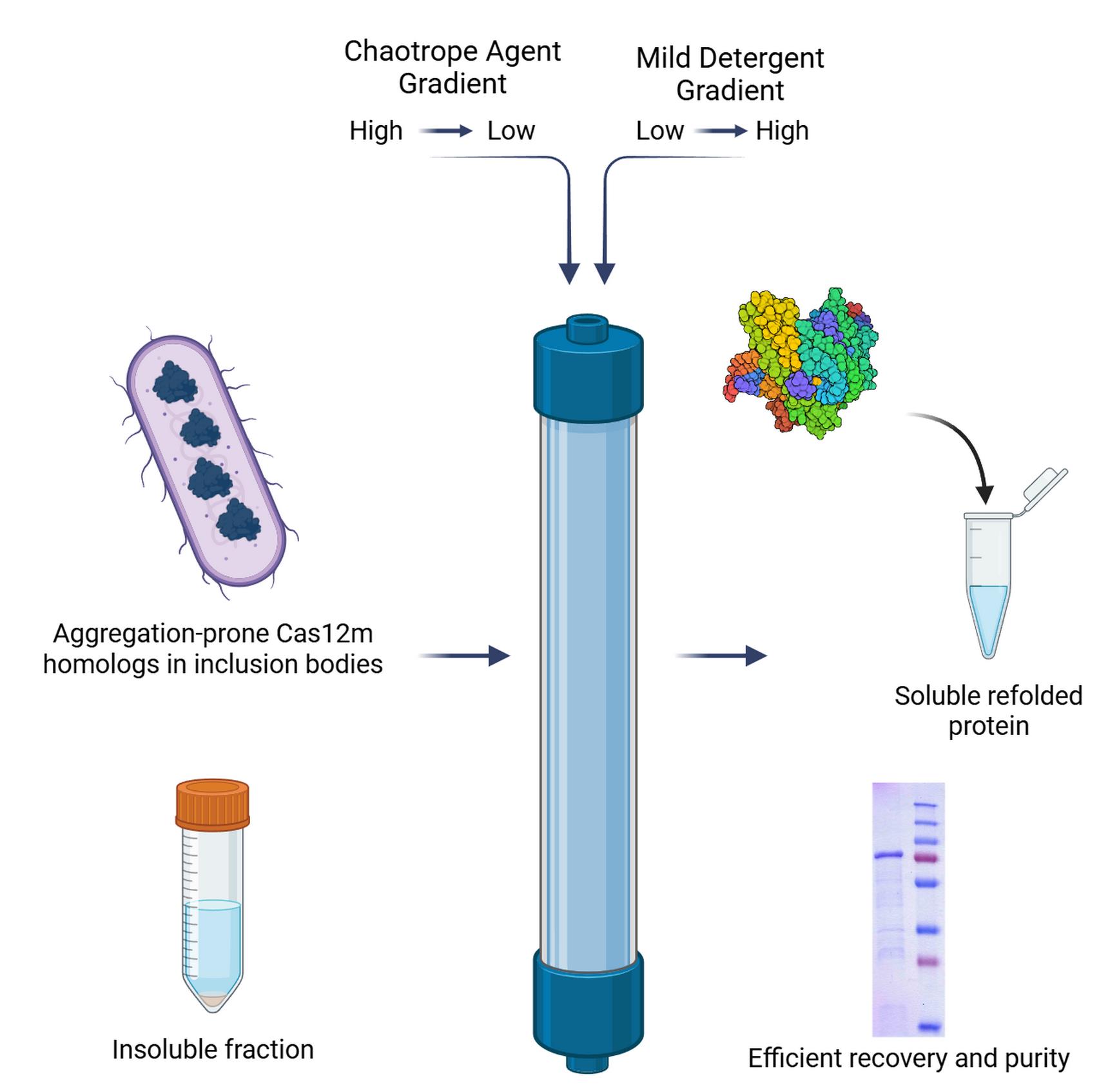
Background
The production of recombinant proteins in heterologous expression systems is a cornerstone of modern molecular biology, biochemistry, and biotechnology. Among the available platforms, Escherichia coli remains one of the most widely used due to its simplicity, cost-effectiveness, and high yield of the target product [1]. However, a major limitation of this system is the frequent formation of inclusion bodies (IBs), insoluble aggregates of misfolded protein [2]. This is particularly common for complex proteins, such as those containing disulfide bonds, multiple subunits, or prosthetic groups, as the intracellular environment of E. coli often cannot support their correct folding [3,4]. While IBs can offer protection from proteolysis and facilitate initial purification, the target protein must be solubilized, refolded, and purified to regain biological activity—a process that remains a significant bottleneck [5].
Traditional methods for refolding proteins from IBs include dilution and dialysis. These techniques rely on reducing the concentration of chaotropic agents used for solubilization (e.g., urea or guanidine hydrochloride) to allow the protein to regain its native conformation [6,7]. However, these methods often suffer from low efficiency and poor reproducibility, primarily due to protein aggregation at intermediate stages of denaturant removal, which is highly dependent on protein concentration and refolding conditions [8].
Chromatographic refolding strategies have emerged as powerful alternatives to overcome these limitations. By immobilizing the denatured protein on a chromatography resin, molecules are spatially separated, thereby minimizing intermolecular aggregation and favoring correct folding [9]. Ion exchange [10,11] and immobilized metal-affinity chromatography (IMAC) [12–14] have been successfully used for this purpose. A particularly effective approach involves the simultaneous application of two gradients during the refolding process: a decreasing gradient of the chaotropic agent and an increasing gradient of a mild detergent. This dual-gradient method helps to suppress aggregation by shielding hydrophobic patches exposed on the protein surface during the critical refolding phase [5,15].
This protocol describes an efficient on-column refolding method using IMAC with a dual-gradient system, developed for a homolog of the Cas12m protein representing subtype CRISPR-CasVM effectors (Dataset S1) [16–18]. Cas12m proteins form binary complexes with crRNA that are capable of binding to DNA with a 5'-TTN-3' PAM but are unable to hydrolyze DNA. The commonly utilized protocol for Cas12m isolation (often with modifications) describes a purification procedure for non-aggregated, soluble proteins [19]. The homolog studied in this work consistently formed inclusion bodies in E. coli across various expression strains and conditions, and traditional refolding methods proved ineffective. The present protocol, utilizing Ni-TED resin and a sodium lauroyl sarcosinate/Tween-20 gradient, enabled the successful production of soluble, albeit partially pure, protein suitable for downstream functional studies. The choice of Ni-TED is due to its high resistance to reducing agents, chelators, and alkaline conditions, which is critical for working with denaturing buffers. Furthermore, the capacity of this resin is lower than that of Ni-NTA (one Ni ion binds only one protein molecule), providing more specific binding compared to polydentate Ni-NTA, reducing nonspecific interactions and increasing the purity of the eluate. The detergent combination was optimized empirically: lauroyl sarcosinate effectively solubilizes inclusion bodies, while the nonionic Tween-20, being transparent at 280 nm, does not interfere with spectrophotometric protein concentration determination. The key advantages of this method over traditional dilution or dialysis are its enhanced reproducibility, reduced aggregation, and the integration of refolding with an initial purification step. This strategy can be adapted for a wide range of other aggregation-prone recombinant proteins that are resistant to conventional refolding approaches.
Materials and reagents
Biological materials
1. E. coli Rosetta 2 (DE3) pLysS (Novagen, catalog number: 71403)
Reagents
1. LB-Miller broth (Difco, catalog number: 244620)
2. Chloramphenicol (Sigma-Aldrich, catalog number: C0378)
3. Kanamycin sulfate (Sigma-Aldrich, catalog number: 60615)
4. Isopropyl β-D-1-thiogalactopyranoside (IPTG) (Sigma-Aldrich, catalog number: I6758)
5. Tris-base (Fisher BioReagents, catalog number: BP152-500)
6. Sodium chloride (NaCl) (Sigma-Aldrich, catalog number: S9888)
7. TritonTM X-100 (Sigma-Aldrich, catalog number: X100)
8. Benzonase® nuclease (Sigma-Aldrich, catalog number: E1014)
9. Magnesium chloride (MgCl2) (Sigma-Aldrich, catalog number: M8266)
10. β-Mercaptoethanol (β-ME) (Sigma-Aldrich, catalog number: M6250)
11. Imidazole (Sigma-Aldrich, catalog number: I2399)
12. Sodium lauroyl sarcosinate (Sigma-Aldrich, catalog number: 61747)
13. Tween® 20 (Sigma-Aldrich, catalog number: P9416)
14. Glycerol (Sigma-Aldrich, catalog number: G5516)
15. Ethylenediaminetetraacetic acid (EDTA) (Sigma-Aldrich, catalog number: E9884)
16. Ethanol, 96% (Sigma-Aldrich, catalog number: 1.00971)
17. Acetic acid, glacial (Sigma-Aldrich, catalog number: A6283)
18. Coomassie Brilliant Blue R-250 (Thermo Scientific, catalog number: 20278)
19. TEMED (Sigma-Aldrich, catalog number: T9281)
20. Sodium dodecyl sulfate (SDS) (Sigma-Aldrich, catalog number: L3771)
21. Ammonium persulfate (APS) (Sigma-Aldrich, catalog number: A3678)
22. Phenylmethylsulfonyl fluoride (PMSF) (Sigma-Aldrich, catalog number: 78830)
23. Prestained protein ladder PageRulerTM Plus, 10–250 kDa (Thermo Scientific, catalog number: 26619)
24. DL-Dithiothreitol (DTT) (Sigma-Aldrich, catalog number: 43815)
25. Ni Seplife FF (TED) chromatography resin (Sunresin New Materials Co Ltd., catalog number: 20211218101)
26. Di-sodium hydrogen phosphate dihydrate (Na2HPO4·2H2O) (Himedia, catalog number: GRM1257)
27. Sodium acetate trihydrate (CH3COONa·3H2O) (Sigma-Aldrich, catalog number: 236500)
28. Sodium hydroxide (NaOH) (Sigma-Aldrich, catalog number: 795429)
29. Acrylamide (Sigma-Aldrich, catalog number: A8887)
30. N,N′-Methylenebisacrylamide (Sigma-Aldrich, catalog number: M7279)
Solutions
1. LB-Miller medium (see Recipes)
2. 1 M MgCl2 (see Recipes)
3. 1 M Tris-base, pH 8.0 (see Recipes)
4. 5 M NaCl (see Recipes)
5. Buffer 1 (see Recipes)
6. Buffer 2 (see Recipes)
7. Buffer 3 (see Recipes)
8. Buffer 4 (see Recipes)
9. Buffer 5 (see Recipes)
10. Buffer 6 (see Recipes)
11. Fixing solution (see Recipes)
12. 12% SDS-PAGE (see Recipes)
13. 50% (v/v) glycerol (see Recipes)
14. Kanamycin (50 mg/mL) (see Recipes)
15. Chloramphenicol (34 mg/mL) (see Recipes)
16. 0.5 M IPTG (see Recipes)
17. Coomassie brilliant blue stain (see Recipes)
18. 0.1 M EDTA, pH 8.0 (see Recipes)
19. 1 M imidazole pH 8.0 (see Recipes)
20. 30% Acrylamide/Bis solution (see Recipes)
21. 1.5 M Tris-Base, pH 8.8 (see Recipes)
22. 0.5 M Tris-Base, pH 6.8 (see Recipes)
23. 10% SDS (see Recipes)
24. 10% APS (see Recipes)
25. 1 M DTT (see Recipes)
26. 10% Triton X-100 solution (see Recipes)
27. 1 M NaOH (see Recipes)
28. Buffer 7 (see Recipes)
29. Buffer 8 (see Recipes)
Recipes
1. LB-Miller medium
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| LB-Miller broth powder | 25 g/L | 25 g |
| Milli-Q water | n/a | to 1 L |
Dissolve 25 g of LB-Miller broth powder in approximately 800 mL of Milli-Q water. Mix thoroughly until all components are completely dissolved. Adjust the final volume to 1 L with Milli-Q water. Sterilize by autoclaving at 121 °C for 20 min. Store the sterilized medium at room temperature.
2. 1 M MgCl2
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| MgCl2·6H2O | 1 M | 101.65 g |
| Milli-Q water | n/a | to 500 mL |
Dissolve 101.65 g of MgCl2·6H2O in 400 mL of Milli-Q water. Once fully dissolved, bring the final volume to 500 mL with Milli-Q water. Filter the buffer using a 0.22 μm filter.
3. 1 M Tris-base, pH 8.0
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Tris-base | 1 M | 121.14 g |
| Milli-Q water | n/a | to 1 L |
Dissolve 121.14 g of Tris-base in approximately 800–900 mL of Milli-Q water with vigorous stirring. Once completely dissolved, adjust the final volume to 1 L with Milli-Q water. Filter the solution through a 0.22 μm filter. The pH of the resulting solution is approximately 10.5 and must be adjusted to pH 8.0 using concentrated HCl before use for most applications.
4. 5 M NaCl
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| NaCl | 5 M | 292.2 g |
| Milli-Q water | n/a | to 1 L |
Dissolve 292.2 g of NaCl in approximately 800 mL of Milli-Q water with stirring. Once completely dissolved, bring the final volume to 1 L with Milli-Q water. Filter the solution through a 0.22 μm filter. Store at room temperature.
5. Buffer 1
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| 1 M Tris-base, pH 8.0 | 40 mM | 40 mL |
| 5 M NaCl | 400 mM | 80 mL |
| 10% Triton X-100 | 0.2% | 2 mL |
| Milli-Q water | n/a | to 1 L |
Add approximately 800 mL of Milli-Q water to a 1 L graduated cylinder or beaker. Add 80 mL of 5 M NaCl stock solution, 40 mL of 1 M Tris-base stock solution (pH 8.0), and 2 mL of 10% Triton X-100 solution. Mix thoroughly until all components are completely dissolved and the solution appears homogeneous. Adjust the final volume to 1 L with Milli-Q water, verify that the pH is 7.8, and adjust if necessary using diluted HCl or NaOH. Filter the buffer through a 0.22 μm filter and store at 4 °C for up to 1 month.
6. Buffer 2
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| 1 M Tris-base, pH 8.0 | 40 mM | 40 mL |
| 5 M NaCl | 400 mM | 80 mL |
| Milli-Q water | n/a | to 1 L |
Add approximately 800 mL of Milli-Q water to a 1 L graduated cylinder or beaker. Add 80 mL of 5 M NaCl stock solution and 40 mL of 1 M Tris-base stock solution (pH 8.0). Mix thoroughly until the solution is homogeneous. Adjust the final volume to 1 L with Milli-Q water, verify that the pH is 7.8, and adjust if necessary using diluted HCl or NaOH. Filter the buffer through a 0.22 μm filter and store at 4 °C for up to 6 months.
7. Buffer 3
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| 1 M Tris-base, pH 8.0 | 40 mM | 40 mL |
| 5 M NaCl | 400 mM | 80 mL |
| 1 M imidazole | 10 mM | 10 mL |
| Sodium lauroyl sarcosinate | 1% (w/v) | 10 g |
| β-ME | 10 mM | 0.714 mL |
| Milli-Q water | n/a | to 1 L |
Add approximately 700 mL of Milli-Q water to a 1 L graduated cylinder or beaker. Add 80 mL of 5 M NaCl stock solution, 40 mL of 1 M Tris-base stock solution (pH 8.0), 10 mL of 1 M imidazole stock solution, and 10 g of sodium lauroyl sarcosinate powder. Mix thoroughly until all components, particularly the detergent powder, are completely dissolved. Add 1 mL of β-ME and mix well. Adjust the final volume to 1 L with Milli-Q water, verify that the pH is 7.8, and adjust if necessary using diluted HCl or NaOH. Filter the buffer through a 0.22 μm filter and store at 4 °C for up to 1 month.
Note: Due to the volatility and oxidation of β-ME, it is recommended to add this component fresh immediately before use if the buffer is stored for more than a few days. The chaotropic agent in this buffer can be optimized (urea 4–8 M or guanidine hydrochloride 4–6 M).
8. Buffer 4
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| 1 M Tris-base, pH 8.0 | 40 mM | 40 mL |
| 5 M NaCl | 400 mM | 80 mL |
| 1 M imidazole, pH 8.0 | 10 mM | 10 mL |
| Tween-20 | 0.1% (v/v) | 1 mL |
| β-ME | 10 mM | 0.714 mL |
| Glycerol (100%) | 5% (v/v) | 50 mL |
| Milli-Q water | n/a | to 1 L |
Add approximately 700 mL of Milli-Q water to a 1 L graduated cylinder or beaker. Add 80 mL of 5 M NaCl stock solution, 40 mL of 1 M Tris-base stock solution (pH 8.0), and 10 mL of 1 M imidazole stock solution. Mix thoroughly until all components are completely dissolved. Add 1 mL of β-ME and 1 mL of Tween-20, then mix well to ensure complete homogeneity. Adjust the final volume to 1 L with Milli-Q water, verify that the pH is 7.8, and adjust if necessary using diluted HCl or NaOH. Filter the buffer through a 0.22 μm filter and store at 4 °C for up to 1 month.
Note: Due to the volatility and oxidation of β-ME, it is recommended to add this component fresh immediately before use if the buffer is stored for more than a few days.
9. Buffer 5
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| 1 M Tris-base, pH 8.0 | 40 mM | 40 mL |
| 5 M NaCl | 400 mM | 80 mL |
| 1 M imidazole, pH 8.0 | 300 mM | 300 mL |
| Glycerol (100%) | 5% (v/v) | 50 mL |
| Tween-20 | 0.1% (v/v) | 1 mL |
| β-ME | 5 mM | 0.357 mL |
| Milli-Q water | n/a | to 1 L |
Add approximately 400 mL of Milli-Q water to a 1 L graduated cylinder or beaker. Add 80 mL of 5 M NaCl stock solution, 40 mL of 1 M Tris-base stock solution (pH 8.0), 300 mL of 1 M imidazole stock solution, and 50 mL of glycerol. Mix thoroughly until the solution is homogeneous. Add 1 mL of Tween-20 and 5 mL of β-ME, then mix well to ensure complete homogeneity. Adjust the final volume to 1 L with Milli-Q water, verify that the pH is 7.8, and adjust if necessary using diluted HCl or NaOH. Filter the buffer through a 0.22 μm filter and store at 4 °C for up to 1 month.
Note: Due to the volatility and oxidation of β-ME, it is recommended to add this component fresh immediately before use if the buffer is stored for more than a few days.
10. Buffer 6
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| 1 M Tris-base, pH 8.0 | 40 mM | 40 mL |
| 5 M NaCl | 400 mM | 80 mL |
| 0.1 M EDTA | 5 mM | 50 mL |
| Glycerol (100%) | 5% (v/v) | 50 mL |
| Tween-20 | 0.1% (v/v) | 1 mL |
| β-ME | 5 mM | 0.357 mL |
| Milli-Q water | n/a | to 1 L |
Add approximately 500 mL of Milli-Q water to a 1 L graduated cylinder or beaker. Add 80 mL of 5 M NaCl stock solution, 40 mL of 1 M Tris-base stock solution (pH 8.0), 50 mL of 0.1 M EDTA stock solution, and 50 mL of glycerol. Mix thoroughly until the solution is homogeneous. Add 1 mL of Tween-20 and 0.35 mL of β-ME, then mix well to ensure complete homogeneity. Adjust the final volume to 1 L with Milli-Q water, verify that the pH is 7.8, and adjust if necessary using diluted HCl or NaOH. Filter the buffer through a 0.22 μm filter and store at 4 °C for up to 1 month.
Note: Due to the volatility and oxidation of β-ME, it is recommended to add this component fresh immediately before use if the buffer is stored for more than a few days.
11. Fixing solution
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Ethanol (96%) | 40% (v/v) | 416.7 mL |
| Glacial acetic acid | 10% (v/v) | 100 mL |
| Milli-Q water | n/a | to 1 L |
Add approximately 300 mL of Milli-Q water to a 1 L graduated cylinder placed in a well-ventilated area or fume hood. Carefully add 416.7 mL of 96% ethanol and 100 mL of glacial acetic acid. Mix thoroughly until the solution is homogeneous. Adjust the final volume to 1 L with Milli-Q water. Store at room temperature in a tightly sealed container to prevent evaporation.
Caution: Prepare this solution in a well-ventilated area or fume hood due to the volatile and corrosive nature of glacial acetic acid. Wear appropriate personal protective equipment.
12. 12% SDS-PAGE (for 2 mini-gels)
a. Resolving gel (10 mL)
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| 30% acrylamide/bis solution | 12% | 4.0 mL |
| 1.5 M Tris-base, pH 8.8 | 0.57 M | 3.8 mL |
| 10% SDS | 0.15% | 150 μL |
| 10% APS | 0.15% | 150 μL |
| TEMED | 0.06% | 6 μL |
| Milli-Q water | n/a | 2.9 mL |
Combine 4.0 mL of 30% acrylamide/bis solution, 3.8 mL of 1.5 M Tris-base (pH 8.8), 150 μL of 10% SDS, 2.9 mL of Milli-Q water, and 150 μL of 10% APS in a 15 mL conical tube. Mix gently by inversion. Immediately before pouring, add 6 μL of TEMED, mix gently but thoroughly, and pour the solution immediately into the gel cassette. Leave appropriate space for the stacking gel and carefully overlay with isopropanol or water to ensure a flat interface. Allow to polymerize completely (approximately 15–30 min).
Caution: Acrylamide is a neurotoxin. Wear appropriate personal protective equipment when handling unpolymerized solutions.
b. Stacking gel (4 mL)
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| 30% acrylamide/bis solution | 5% | 670 μL |
| 0.5 M Tris-base, pH 6.8 | 0.125 M | 1.0 mL |
| 10% SDS | 0.1% | 40 μL |
| 10% APS | 0.1% | 40 μL |
| TEMED | 0.1% | 4 μL |
| Milli-Q water | n/a | 2.25 mL |
After polymerization of the resolving gel, remove the overlay and rinse the gel surface with Milli-Q water. Combine 670 μL of 30% acrylamide/bis solution, 1.0 mL of 0.5 M Tris-base (pH 6.8), 40 μL of 10% SDS, 2.25 mL of Milli-Q water, and 40 μL of 10% APS in a 15 mL conical tube. Mix gently by inversion. Add 4 μL of TEMED, mix gently but thoroughly, and pour immediately on top of the polymerized resolving gel. Immediately insert a clean comb, avoiding air bubbles. Allow to polymerize completely (approximately 10–15 min).
Caution: Acrylamide is a neurotoxin. Wear appropriate personal protective equipment when handling unpolymerized solutions.
13. 50% (v/v) glycerol
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Molecular biology–grade glycerol | 50% (v/v) | 50 mL |
| Milli-Q water | n/a | 50 mL |
Add 50 mL of molecular biology–grade glycerol to 50 mL of Milli-Q water in a suitable container. Mix thoroughly on a magnetic stirrer until the solution is completely homogeneous. Store at room temperature.
14. Kanamycin (50 mg/mL)
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Kanamycin sulfate | 50 mg/mL | 2.5g |
| Sterile Milli-Q water | n/a | to 50 mL |
Dissolve 2.5 g of kanamycin sulfate in approximately 40 mL of sterile Milli-Q water. After complete dissolution, adjust the final volume to 50 mL with sterile Milli-Q water. Sterilize the solution by filtration through a 0.22 μm filter. Aliquot into sterile tubes and store at -20 °C.
15. Chloramphenicol (34 mg/mL)
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Chloramphenicol | 34 mg/mL | 1.7 g |
| Absolute ethanol | 100% (v/v) | 50 mL |
Dissolve 1.7 g of chloramphenicol in 50 mL of absolute ethanol. Mix thoroughly until completely dissolved. The solution is sterile and does not require filtration. Aliquot and store at -20 °C protected from light.
16. 0.5 M IPTG
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| IPTG | 0.5 M | 5.95 g |
| Milli-Q water | n/a | to 50 mL |
Dissolve 5.95 g of IPTG in approximately 40 mL of Milli-Q water. After complete dissolution, adjust the final volume to 50 mL with Milli-Q water. Sterilize the solution by filtration through a 0.22 μm filter. Aliquot into sterile tubes and store at -20 °C.
17. Coomassie Brilliant Blue staining solution
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Coomassie Brilliant Blue R-250 | 0.1% (w/v) | 1.0 g |
| Ethanol (absolute) | 45% (v/v) | 450 mL |
| Glacial acetic acid | 10% (v/v) | 100 mL |
| Milli-Q water | n/a | 450 mL |
Dissolve 1.0 g of Coomassie Brilliant Blue R-250 in a mixture of 450 mL of absolute ethanol and 100 mL of glacial acetic acid, using a magnetic stirrer until the dye is completely dissolved. Add 450 mL of Milli-Q water and mix thoroughly until homogeneous. Filter the solution through filter paper before use to remove any undissolved particles. Store at room temperature in a tightly sealed container.
Caution: Glacial acetic acid is corrosive and volatile. Prepare this solution in a well-ventilated area or fume hood and wear appropriate personal protective equipment.
18. 0.1 M EDTA, pH 8.0
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| EDTA disodium salt dihydrate | 0.1 M | 37.22 g |
| Milli-Q water | n/a | to 1 L |
Dissolve 37.22 g of EDTA disodium salt dihydrate in approximately 800 mL of Milli-Q water while stirring vigorously. Adjust the pH to 8.0 using NaOH pellets or a concentrated (e.g., 1 M) NaOH solution; note that the EDTA will not fully dissolve until the pH approaches 8.0. Once completely dissolved and at the correct pH, adjust the final volume to 1 L with Milli-Q water. The solution can be sterilized by autoclaving at 121 °C for 20 min if required. Store at room temperature.
19. 1 M imidazole, pH 8.0
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Imidazole | 1 M | 34.04 g |
| Milli-Q water | n/a | to 500 mL |
Dissolve 34.04 g of imidazole in approximately 400 mL of Milli-Q water. While stirring, adjust the pH to 8.0 using concentrated HCl. Once the pH is stable and the imidazole is completely dissolved, adjust the final volume to 500 mL with Milli-Q water. Filter-sterilize the solution through a 0.22 μm filter and store at room temperature.
20. 30% acrylamide/bis solution
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Acrylamide | 29.2% (w/v) | 146 g |
| N, N'-methylenebisacrylamide | 0.8% (w/v) | 4 g |
| Milli-Q water | n/a | to 500 mL |
Dissolve 146 g of acrylamide and 4 g of N, N'-methylenebisacrylamide in approximately 400 mL of Milli-Q water with gentle stirring. Once completely dissolved, adjust the final volume to 500 mL with Milli-Q water. Filter the solution through a 0.45 μm filter. Store in a dark bottle or container wrapped in aluminum foil at 4 °C to protect from light.
Caution: Acrylamide is a potent neurotoxin and a suspected carcinogen. Always wear appropriate personal protective equipment (gloves, lab coat, safety glasses) when handling unpolymerized acrylamide. Avoid inhalation of powder and skin contact.
21. 1.5 M Tris-base, pH 8.8
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Tris-base | 1.5 M | 90.85 g |
| Milli-Q water | n/a | to 500 mL |
Dissolve 90.85 g of Tris-base in approximately 400 mL of Milli-Q water. While stirring, adjust the pH to 8.8 using concentrated HCl. Note that a significant volume of HCl may be required, and the solution will generate heat during neutralization. Allow the solution to cool to room temperature after pH adjustment, then check and adjust the pH again if necessary. Once the pH is stable at 8.8 and the Tris-base is completely dissolved, adjust the final volume to 500 mL with Milli-Q water. The solution can be sterilized by autoclaving at 121 °C for 20 min if required. Store at room temperature.
22. 0.5 M Tris-base, pH 6.8
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Tris-base | 0.5 M | 30.3 g |
| Milli-Q water | n/a | to 500 mL |
Dissolve 30.3 g of Tris-base in approximately 400 mL of Milli-Q water. While stirring, carefully adjust the pH to 6.8 using concentrated HCl. The solution will generate heat during neutralization; allow it to cool to room temperature before making final pH adjustments. Once the pH is stable at 6.8 and the Tris-base is completely dissolved, adjust the final volume to 500 mL with Milli-Q water. The solution can be sterilized by autoclaving at 121 °C for 20 min if required. Store at room temperature.
23. 10% SDS
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| SDS | 10% (w/v) | 50 g |
| Milli-Q water | n/a | to 500 mL |
Dissolve 50 g of SDS in approximately 400 mL of Milli-Q water with gentle stirring and heating to 50 °C to facilitate dissolution. Once fully dissolved and when the solution appears clear, adjust the final volume to 500 mL with Milli-Q water. Store at room temperature.
Caution: Wear appropriate personal protective equipment, including a mask, when handling SDS powder to avoid inhalation.
24. 10% APS
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| APS | 10% (w/v) | 1 g |
| Milli-Q water | n/a | 10 mL |
Dissolve 1 g of APS in 10 mL of Milli-Q water. Mix thoroughly until completely dissolved. Aliquot into small volumes (e.g., 0.5–1 mL) and store at -20 °C.
Note: The solution is stable for several weeks when stored frozen. For best results in polymerization reactions, use freshly prepared solution or aliquots that have undergone minimal freeze-thaw cycles.
25. 1 M DTT
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| DTT | 1 M | 1.54 g |
| Milli-Q water | n/a | to 10 mL |
Dissolve 1.54 g of DTT in approximately 8 mL of Milli-Q water. Once fully dissolved, adjust the final volume to 10 mL with Milli-Q water. Sterilize the solution by filtration through a 0.22 μm filter. Aliquot into small working volumes and store at -20 °C.
Note: DTT solutions are susceptible to oxidation. For critical applications, prepare fresh solutions or use aliquots that have undergone minimal freeze-thaw cycles.
26. 10% Triton X-100 solution
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Triton X-100 | 10% (v/v) | 10 mL |
| Milli-Q water | n/a | to 100 mL |
Add 10 mL of Triton X-100 to approximately 80 mL of Milli-Q water. Mix thoroughly using a magnetic stirrer until the solution becomes clear and homogeneous; this may take several minutes. Adjust the final volume to 100 mL with Milli-Q water. Store at 4 °C.
27. 1 M NaOH
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| NaOH | 1 M | 4 g |
| Milli-Q water | n/a | to 100 mL |
Measure approximately 80 mL of water into a volumetric flask or beaker.
Caution: Add 4.0 g of NaOH while stirring. Dissolution is highly exothermic; the solution will become hot. Allow the solution to cool to room temperature. Bring the volume up to the 100 mL mark with water. Mix thoroughly.
28. Buffer 7
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Na2HPO4·2H2O | 0.4 M | 7.1 g |
| NaCl | 0.15 M | 0.88 g |
| Milli-Q water | n/a | to 100 mL |
Measure approximately 80 mL of water into a volumetric flask. Dissolve 7.1 g of Na2HPO4·2H2O in it. Add 0.88 g of NaCl and stir until completely dissolved. Check the pH of the solution. The Na2HPO4 solution will have an alkaline pH (usually ~8.9–9.0). Slowly add concentrated HCl dropwise while stirring and monitoring with a pH meter, until the pH reaches exactly 7.0.
Tip: It is better to dilute a small portion of HCl (e.g., to 1 M) for more precise adjustment.
Bring the volume up to the 100 mL mark with water.
29. Buffer 8
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| CH3COONa·3H2O | 0.4 M | 0.14 g |
| Ethanol | 20% | 20 mL |
| Milli-Q water | n/a | to 100 mL |
Prepare 10 mM acetate buffer, pH 6.5: Dissolve 1.36 g of CH3COONa·3H2O in approximately 20 mL of water. Measure the pH. The sodium acetate solution will have an alkaline pH. Slowly, while stirring and monitoring with a pH meter, add acetic acid until the pH becomes 6.5. Bring the volume of this buffer up to 80 mL with water. Mix well. To 80 mL of the finished acetate buffer (pH 6.5), add 20 mL of ethanol. Mix thoroughly.
Laboratory supplies
1. Safe-lock tubes, 0.5 mL (Eppendorf, catalog number: 0030121023)
2. Safe-lock tubes, 1.5 mL (Eppendorf, catalog number: 0030120086)
3. Safe-lock tubes, 2.0 mL (Eppendorf, catalog number: 0030120094)
4. Falcon conical centrifuge tubes, 15 mL (Corning, catalog number: 352095)
5. Falcon conical centrifuge tubes, 50 mL (Corning, catalog number: 352070)
6. Dialysis tubing SnakeSkin, 10 K MWCO (Thermo Scientific, catalog number: 68100)
7. Centrifugal filters JetSpin, 10 KDa MWCO (JetBioFil, catalog number: FTT410500)
8. Serological pipettes, 10 mL (GenFollower, catalog number: YSPS10-1)
9. Petri dishes (Sigma-Aldrich, catalog number: P5481-500EA)
10. Pipette tips (various volumes) (Eppendorf, catalog numbers: EP0030000870-1PAK, EP0030078578-960EA, EP0030078519-960EA)
11. Volumetric flasks (Glassco, catalog numbers: Q138.510.01, Q138.510.04, Q138.510.06)
12. Beakers (MiniMed, catalog numbers: В-1-150, В-1-250, Н-1-600)
13. Erlenmeyer flasks (HARIO, catalog number: SF-1L SCI)
14. Calibrated pipettes (various volumes) (DLAB TopPette, catalog number: 7010101014, LH-7290207010101009)
Equipment
1. Water bath thermostat ELMI TW-2 (ELMI, catalog number: TW-2)
2. Standard incubator binder BD 56 (Binder, catalog number: 9010-0323)
3. Biological safety cabinet Labconco Purifier Logic+ Class II, Type A2 (Labconco, catalog number: 302481101)
4. Stackable incubator shaker Innova® S44i (Eppendorf, catalog number: S44I300001)
5. Spectrophotometer SmartSpec Plus (Bio-Rad, catalog number: 170-2525)
6. Portable mini centrifuge Eppendorf MiniSpin (Eppendorf, catalog number: 5452000010)
7. High-speed centrifuge Beckman Coulter Avanti J-E (Beckman Coulter, catalog number: 369001)
8. High-speed benchtop centrifuge Eppendorf Centrifuge 5804R (Eppendorf, catalog number: 5805000010)
9. Digital sonifier Branson 450 (Branson, catalog number: 100-132-889R)
10. Empty chromatography column 5 mL (Smart-Lifesciences, catalog number: SLM011)
11. Chromatography system ÄKTA Pure (Cytiva, catalog number: 29018226)
12. Magnetic stirrer Heidolph MR Hei-Standard (Heidolph, catalog number: 505-20000-00)
13. Vertical polyacrylamide gel electrophoresis system Bio-Rad Mini-PROTEAN Tetra Cell (Bio-Rad, catalog number: 1658005EDU)
14. Electrophoresis power supply Bio-Rad PowerPac Basic (Bio-Rad, catalog number: 1645050)
15. Thermo-shaker with cooling BioSan TS-100C (BioSan, catalog number: BS-010143-AAI)
16. Spectrophotometer Thermo Fisher Scientific NanoDrop 1000 (Thermo Fisher Scientific, catalog number: ND1000)
17. Ultra-low temperature freezer Sanyo MDF-U53V (Sanyo, catalog number: SM9910073)
18. Water purification system Rephile NuZar Q (Rephile, catalog number: RN0Q00000K)
19. pH-meter Mettler Toledo S20 SevenEasyTM (Mettler Toledo, catalog number: S20)
20. High-pressure steam sterilizer (autoclave) Sanyo MLS-3781L (Sanyo, catalog number: SM6610004)
21. Lab balance Mettler Toledo XPE204 (Mettler Toledo, catalog number: 30087643)
22. Sterile disposable filter unit Nalgene Rapid-Flow (Thermo Fisher Scientific, catalog number: 566-0020)
23. Freezer (-20 °C) ХЛ-250 POZIS (Pozis, catalog number: 223TV)
24. Refrigerator (2–8 °C) ХЛ-250 POZIS (Pozis, catalog number: 223TV)
Software and datasets
1. UNICORN 7 control software (Cytiva)
2. Image Lab Software (Bio-Rad)
Procedure
A. Expression of recombinant proteins
Note: In this specific case, the E. coli strain Rosetta 2 (DE3)pLysS was selected because it provided the highest yield of the target protein in inclusion bodies. However, for a different protein, the optimal expression strain and conditions (e.g., host strain, induction temperature, and inducer concentration) may vary. Therefore, before proceeding with this purification protocol, users should perform their own small-scale expression tests to confirm that their target protein predominantly forms inclusion bodies and to identify the conditions that maximize its insoluble expression.
1. Transform the verified plasmids into chemically competent E. coli Rosetta 2 (DE3)pLysS cells according to the manufacturer's instructions. After the heat shock, add 1 mL of LB medium and incubate the cells for 1 h at 37 °C and 200 rpm.
2. Spread 100–200 μL of the cell suspension onto an LB agar plate containing the appropriate antibiotics: kanamycin (50 μg/mL) and chloramphenicol (34 μg/mL). Incubate the plate upside down at 37 °C for 16 h (or until colonies appear).
3. Using a sterile loop, pick a single well-formed colony. Inoculate the colony into a separate tube or flask containing 35 mL of LB-Miller medium with the appropriate antibiotics. Incubate the culture for 16 h (overnight) at 37 °C and 200 rpm in a thermo shaker.
4. Prepare 3.5 L of LB-Miller medium in flasks of a suitable volume. Add kanamycin to a final concentration of 50 μg/mL. Inoculate the main culture with the pre-culture at a 1:100 ratio.
5. Incubate the culture at 37 °C with constant shaking (200 rpm), monitoring the optical density (OD600). Continue cultivation until an OD600 of ~0.6 is reached (mid-logarithmic growth phase).
6. Withdraw a 1 mL pre-induction sample (this can be used as a negative control for SDS-PAGE). Add an IPTG solution to the main culture to a final concentration of 0.2 mM.
7. After adding IPTG, continue incubating the culture with constant shaking (200 rpm) for 16 h at 24 °C.
B. Cell lysis and inclusion body isolation
Note: All procedures, unless otherwise specified, should be performed at 4 °C (on ice or in a cold room). Use pre-cooled buffers and centrifuge rotors.
1. Harvest the cells from the 3.5 L culture by centrifugation at 6,000× g for 15 min. Carefully decant and discard the supernatant. The cell pellet can either be used immediately for purification or frozen at -20 °C or -80 °C for long-term storage.
2. Completely resuspend the cell pellet in 100 mL of Buffer 1 (40 mM Tris-base pH 7.8, 400 mМ NaCl, 0.2% Triton-X100). Thorough pipetting or the use of a dispenser may be required to break up the pellet completely.
3. Lyse the cell membranes using an ultrasonic disintegrator with the following settings to minimize heating: amplitude 45%, 2 s pulse-on, 6 s pulse-off, for a total pulse-on time of 300 s. Perform the sonication in a beaker placed on an ice bath. The suspension should become clearer and less viscous.
4. Transfer the lysate into appropriate centrifuge tubes and centrifuge at 28,000× g for 30 min at 4 °C to pellet the insoluble fraction (inclusion bodies).
C. Washing of inclusion bodies
1. Carefully decant and discard the supernatant (contains soluble cellular components). The pellet (inclusion bodies) should be dense.
2. Gently resuspend the pellet in 100 mL of Buffer 2 (40 mM Tris-Base pH 7.8, 400 mM NaCl). Use a pipette, a glass rod, or an ultrasonic disintegrator with amplitude 25%, 2 s pulse-on, 2 s pulse-off, for a total pulse-on time of 300 s, to completely resuspend the pellet.
3. Add MgCl2 to the suspension to a final concentration of 1 mM and 1 μL of Benzonase (or according to the manufacturer's instructions). Incubate the suspension for 30 min at room temperature with constant agitation on a shaker or magnetic stirrer.
4. Pellet the inclusion bodies by centrifugation at 28,000× g for 30 min at 4 °C.
5. Repeat steps C1–4 (resuspension in buffer and centrifugation) two more times to remove residual contaminants. After the final centrifugation, thoroughly remove the supernatant.
D. Solubilization of inclusion bodies
1. Resuspend the purified inclusion bodies in Buffer 3 (40 mM Tris-base pH 7.8, 400 mM NaCl, 10 mM β-Mercaptoethanol, 10 mM imidazole, 1% sodium lauroyl sarcosinate). The volume of Buffer 3 should be determined empirically to completely cover the pellet (e.g., 20–50 mL).
2. For complete dissolution, treat the suspension with ultrasound on ice using a gentle mode: amplitude 25%, 2 s pulse-on, 2 s pulse-off, for a total pulse-on time of 600 s.
3. Clarify the solution by centrifugation at 28,000× g for 30 min at 4 °C. Transfer the clear supernatant to a new tube.
E. Column packing
1. Measure out 10 mL of Ni Seplife FF (TED) resin suspension and mix it thoroughly before use.
2. Place the column vertically in a stand. Slowly pour the resin suspension into the column, allowing the excess liquid to drain out through the column outlet by gravity flow (Figure 1).
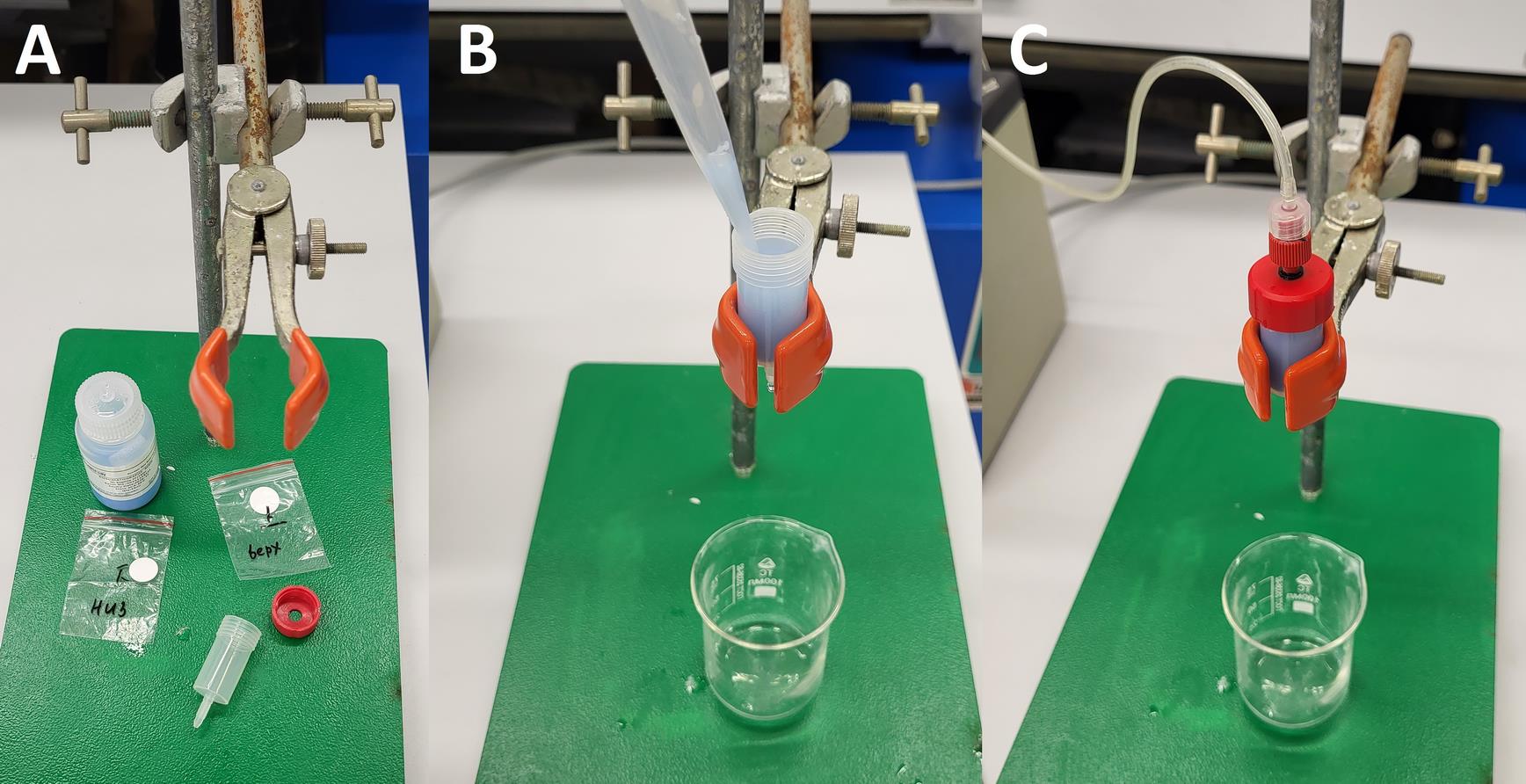
Figure 1. Column packing. (A) Empty chromatography column. (B) Applying resin into the column. (C) Washing column.
3. Carefully insert the top frit and adapter, ensuring they fit evenly across the entire surface. Wash the column with 5–10 column volumes (CV) of Milli-Q water for equilibration; the recommended flow rate is 3 mL/min. Monitor the flow for uniformity and stability of the resin bed.
Note: Perform all operations at room temperature. Avoid sudden movements and pressure fluctuations. Store at 4 °C with the end caps closed. For long-term storage, add a preservative (20% ethanol).
Caution: Specific packing conditions may be required for different types of resins.
F. Affinity chromatography and on-column refolding
1. Equilibrate the column with at least 5–10 CVs of Buffer 3.
2. Slowly load the clear supernatant onto the equilibrated column. The recommended flow rate is 1–2 mL/min. Collect the flowthrough fractions, which contain components that did not bind to the resin.
3. Wash the column with 30 mL (6 CVs) of the starting buffer (Buffer 3) to completely remove any unbound components.
4. Perform refolding of the target protein by gradually decreasing the concentration of sodium lauroyl sarcosinate from 1% to 0%, while simultaneously increasing the concentration of Tween-20 from 0% to 0.1%. To achieve this, use the Gradient function in the UNICORN software to perform a complete buffer exchange from Buffer 3 to Buffer 4 (40 mM Tris-base pH 7.8, 400 mM NaCl, 10 mM β-Mercaptoethanol, 10 mM imidazole, 0.1% Tween-20) over 10 CVs (50 mL) at a flow rate of 0.5 mL/min.
5. After refolding is complete, elute the target protein from the column using the elution buffer (Buffer 5) (40 mM Tris-base pH 7.8, 400 mM NaCl, 300 mM imidazole, 0.1% Tween-20, 5 mM β-ME, 5% glycerol). The recommended flow rate is 1–2 mL/min. Collect fractions of 1–2 mL each (Figure 2).
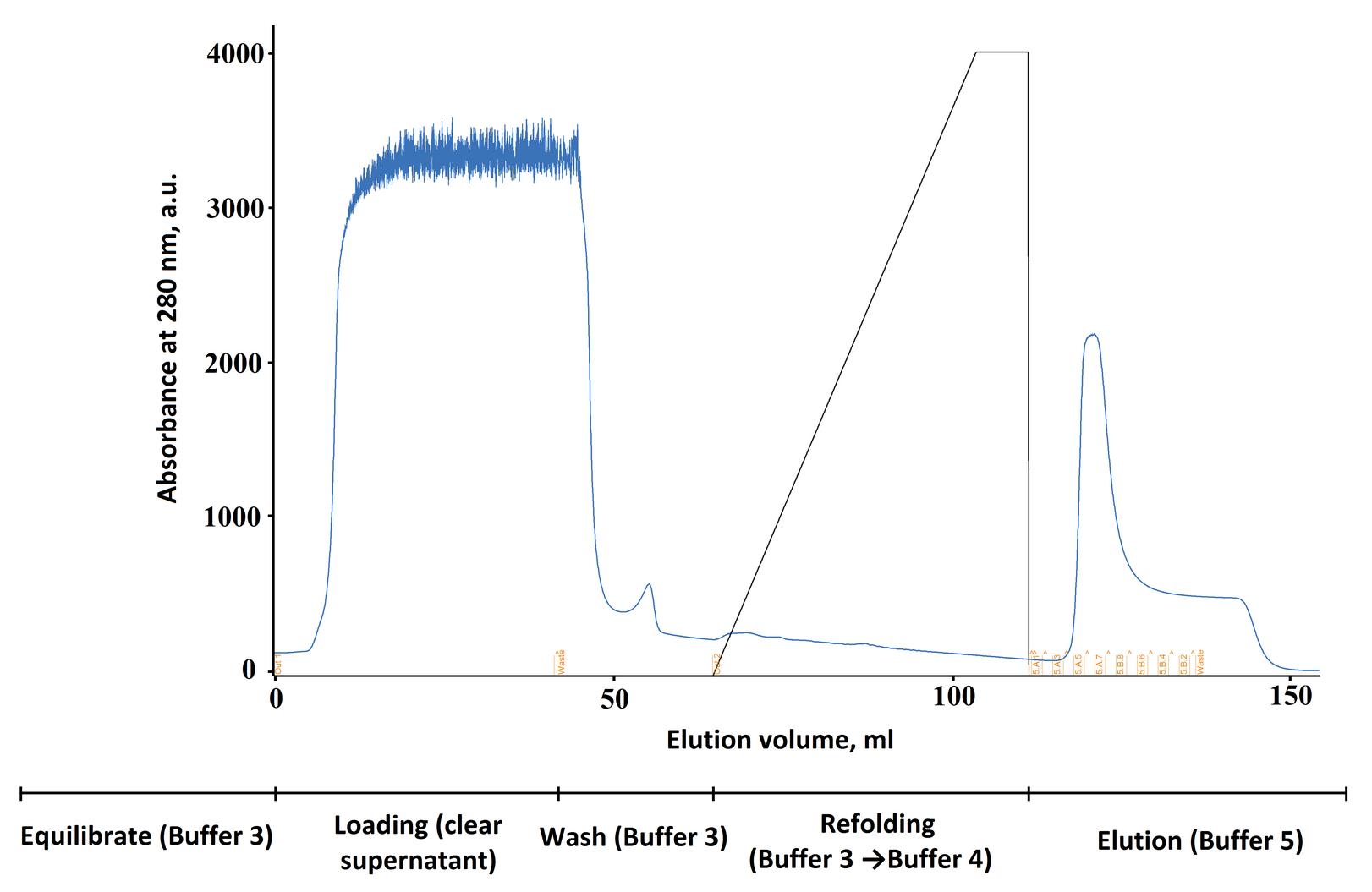
Figure 2. Refolding chromatogram of the target protein on the Ni-TED column. The black line represents the gradient of Buffer 4 in Buffer 3. The timeline below the graph shows the main stages and the buffers used.
6. Analyze the collected fractions using SDS-PAGE (see Section G). Pool the fractions containing the target protein of high purity (in this example, fractions 7–10 in Figure 3).
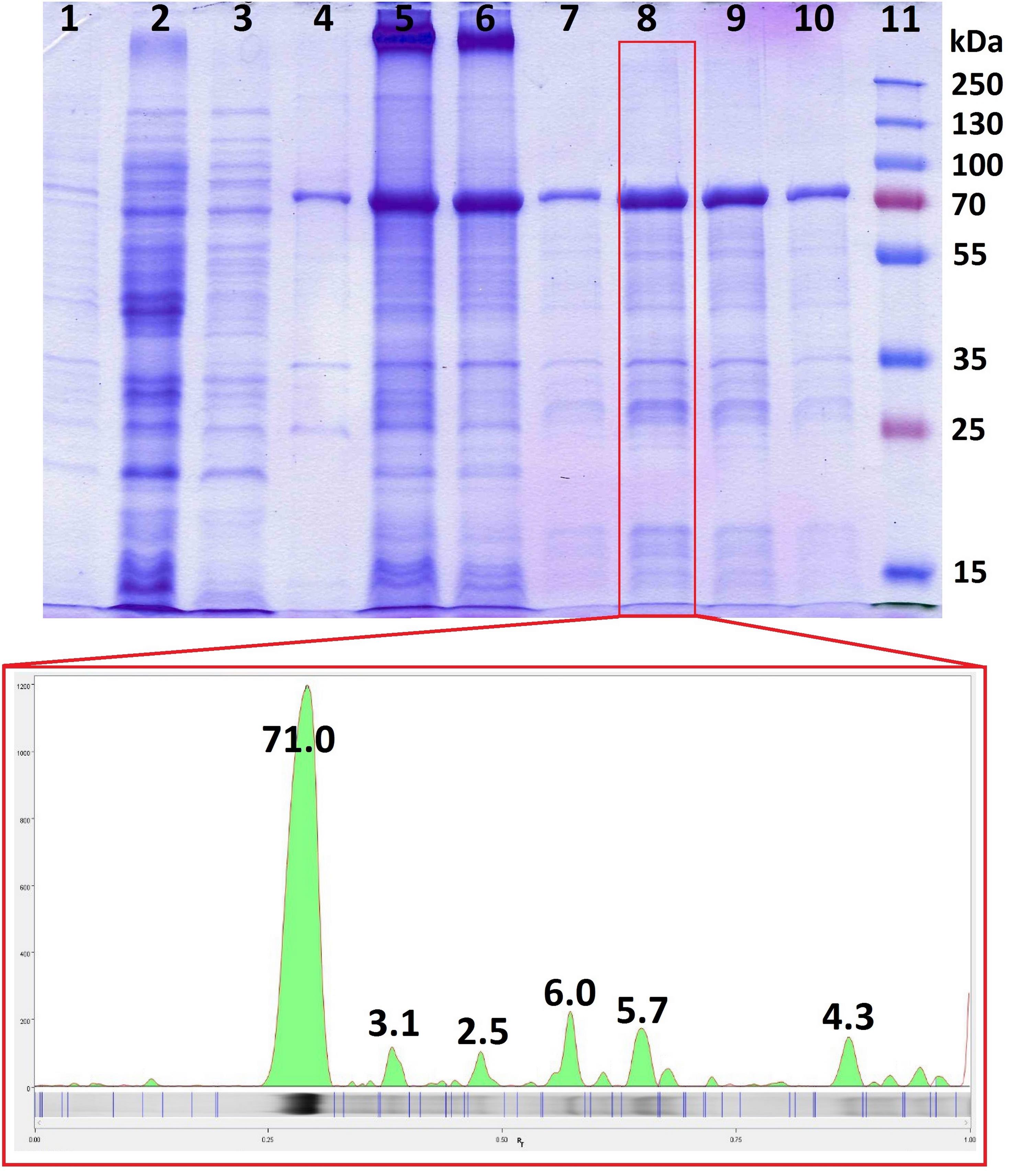
Figure 3. Purity analysis by SDS-PAGE. Quantitative densitometric analysis of band intensity was performed using Image LabTM Software. 1: Total cell lysate. 2: Clarified cell lysate. 3: Supernatant after inclusion body wash. 4: Washed inclusion bodies. 5: Solubilized inclusion bodies. 6: Column flowthrough. 7–10: Column elution fractions. 11: Prestained protein ladder PageRuler Plus, 10–250 kDa.
7. Place the pooled fractions into a dialysis membrane (MWCO 10 kDa) and dialyze against 1 L of Buffer 6 (40 mM Tris-base pH 7.8, 400 mM NaCl, 5 mM EDTA, 0.1% Tween-20, 5 mM β-ME, 5% glycerol) overnight at 4 °C with constant stirring.
8. Transfer the dialyzed protein to a centrifugal concentrator (MWCO 10 kDa). Concentrate the sample to the desired volume, following the manufacturer's instructions. Determine the protein concentration using a suitable method (e.g., Bradford assay). Aliquot the protein and store it at -80 °C.
Note: One could attempt to repeat the procedure; however, in our case, this additional step did not increase the yield of the target protein. We recommend focusing on optimizing the primary protein purification process and discarding the obtained flowthrough fractions.
G. Purity analysis by SDS-PAGE
1. Prepare samples (e.g., 10–20 μg of protein or 10–15 μL from fractions) by mixing them with sample loading buffer, and denature at 95 °C for 5 min.
2. Load the samples into the wells of a 12% polyacrylamide gel.
3. Perform electrophoresis in a chamber filled with Tris-glycine-SDS running buffer at a constant voltage of 120 V until the dye front reaches the bottom of the gel.
4. Place the gel in a fixing solution (40% ethanol, 10% acetic acid) for 5–10 min.
5. Transfer the gel to a 0.1% Coomassie R-250 solution prepared in the same fixative for 10–20 min.
6. Remove excess dye by washing the gel with hot water (50–60 °C) until clear protein bands appear against a transparent background.
E. Regeneration and storage of a chromatography column
1. To remove residual buffer, salts, solvents, or weakly bound substances from the previous chromatographic run, set the flow rate to 2 mL/min. Pass 10 CVs of H2O through the column.
2. For deep cleaning of the matrix from tightly bound biological contaminants and inactivation of microorganisms, pass 10 CVs of 1 M NaOH solution through the column at a flow rate of 2 mL/min.
3. To completely eliminate residual alkali that could disrupt subsequent buffers or damage the column, flush the system with 10 CVs of water at 2 mL/min.
4. Equilibrate the column to a stable, physiological ionic state by passing 10 CVs of Buffer 7 (0.15 M NaCl, 0.4 M Na2HPO4, pH 7.0) at 2 mL/min, ensuring it is properly conditioned for the next run or free from residual acids/alkalis before storage.
5. To prevent salt crystallization and potential column clogging during storage, thoroughly rinse the system by passing 10 CVs of water at 2 mL/min to remove all residual buffer salts.
6. To preserve the column by preventing microbial growth and matrix hydrolysis, pass 10 CVs of storage buffer (Buffer 8) (20% ethanol, 10 mM CH3COONa, pH 6.5) through it at a flow rate of 2 mL/min.
7. Immediately install storage caps on both ends of the column (inlet and outlet). This prevents ethanol evaporation and drying of the matrix, which is the most common cause of irreversible column damage.
Note: Store the column at 4 °C (in a refrigerator). This further suppresses microbial growth and slows any chemical processes. Never freeze the column.
Data analysis
A. Protein purity and yield assessment
1. Analyze all fractions (flowthrough, wash, elution) by SDS-PAGE using 12% polyacrylamide gels.
2. Visualize protein bands by Coomassie Brilliant Blue R-250 staining.
3. Estimate protein purity by densitometric analysis of gel images using Image Lab or similar software.
4. Determine protein concentration using a Bradford assay with bovine serum albumin as the standard.
5. Calculate total protein yield from elution fractions based on concentration measurements.
B. Quality control criteria
1. Include fractions for pooling only if the target protein represents >70% of the total protein by densitometry.
2. Exclude samples showing significant proteolytic degradation or smearing on SDS-PAGE.
3. Accept protein preparations with an A260/A280 ratio < 0.7, indicating minimal nucleic acid contamination.
C. Software requirements
UNICORN software for chromatography data collection and gradient control; Image Lab for gel densitometry analysis; standard spreadsheet software for statistical calculations.
Validation of protocol
Using this protocol, the Cas12m effector protein representing subtype V-M of CRISPR-Cas systems was isolated. After column elution and concentration of the fractions containing the target protein, 6.75 mg were obtained. The optical density ratio OD260/OD280 = 0.66 indicated minimal nucleic acid contamination. The quality of the resulting protein sample is sufficient for studying its RNA- and DNA-binding activity.
The ability of the isolated protein to form the Cas12m-crRNA binary complex and the Cas12m-crRNA-DNA ternary complex was confirmed by electrophoretic mobility shift assay (EMSA) [20]. Binary complexes formation analysis was performed by incubating 0, 0.5, 1, and 1.5 μg of Cas12m protein with crRNA for 1 h at 37 °C in binding buffer (Table 1). Reaction products were fractionated by electrophoresis using 7% TBE-PAGE gel and analyzed on the Gel Doc EZ System (Figure 4A). Ternary complexes formation analysis was performed by incubating 0, 0.5, 1, and 1.5 μg of Cas12m protein with crRNA and target DNA for 1 h at 37 °C in binding buffer. Reaction products were run on a 2% agarose gel in TBE buffer and analyzed on the Gel Doc EZ System (Figure 4B).
Table 1. Binding buffer
| Reagent | Final concentration | Quantity or volume |
| 1 mg/mL protein | 0.025, 0.05, or 0.075 μg/μL | 0.5, 1 μL, or 1.5 μL |
| crRNA | 0.25 μM | Variable* |
| Target | 0.25 μM | Variable** |
| 10× binding buffer | 1× | 2 μL |
| Milli-Q water | n/a | to 20 μL |
*moles ssRNA (mol) = mass of ssRNA (g)/[(length of ssRNA (nt) × 320.47 g/mol) + 18.02 g/mol]
**moles dsDNA (mol) = mass of dsDNA (g)/[(length of dsDNA (bp) × 615.94 g/mol/bp) + 36.04 g/mol]
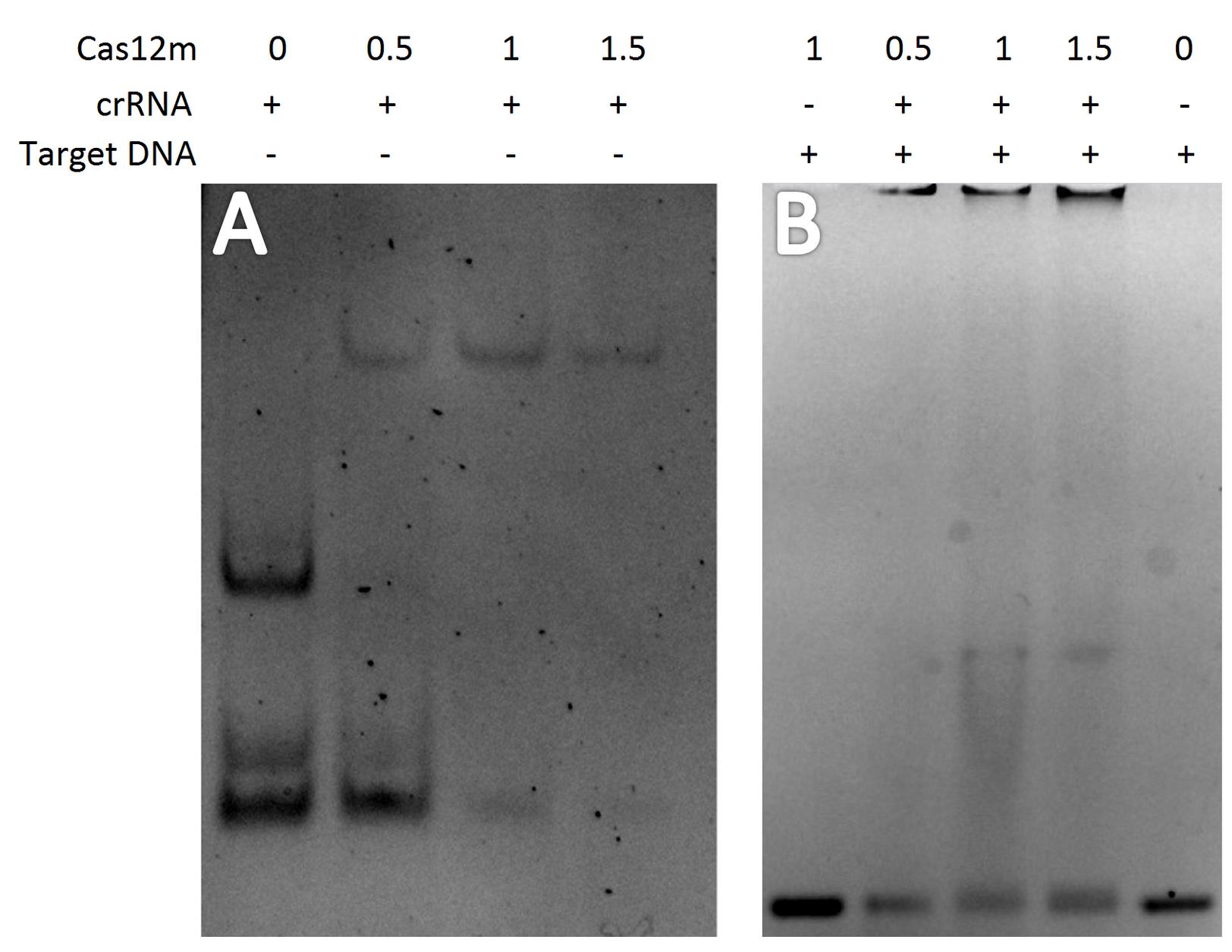
Figure 4. Analysis of Cas12m complex formation. (A) Binary Cas12m-crRNA complexes. (B) Ternary Cas12m-crRNA-DNA complexes.
General notes and troubleshooting
General notes
1. Protein stability and storage
• Refolded proteins should be aliquoted and stored at -80 °C to prevent repeated freeze-thaw cycles.
• For long-term storage, consider adding 5%–10% glycerol as a cryoprotectant.
• Avoid storing proteins at high concentrations (>5 mg/mL) to minimize aggregation.
2. Buffer optimization
• The chaotropic agent in Buffer 3 can be optimized (urea 4–8 M or guanidine hydrochloride 4–6 M).
• Alternative mild detergents (CHAPS, Triton X-114) can be tested in the refolding gradient.
• For proteins requiring specific redox conditions, adjust β-mercaptoethanol concentration (1–10 mM).
3. System applicability
• This protocol can be adapted for proteins with different affinity tags (GST, MBP) using appropriate resins.
4. Scale considerations
• For larger scales (>50 mL resin), increase gradient volumes proportionally (15–20 CVs).
• Maintain constant linear flow rate during the refolding step (0.2–0.5 mL/min).
• Consider bed height to diameter ratio (recommended 5:1 to 10:1) for optimal refolding.
5. Quality control
• Regular column cleaning and sanitization are recommended after each use.
6. Limitations of the method
• The protocol is fundamentally dependent on the presence of a functional affinity 6xHis-tag. Proteins with inaccessible or cleaved tags will not bind efficiently, leading to significant loss.
• While we provide a robust starting point, the optimal chaotropic agent (e.g., urea vs. sarcosinate concentration) and the slope of the refolding gradient may require protein-specific adjustment to maximize the yield of correctly folded protein.
• A significant fraction of the solubilized protein often does not bind to the Ni-TED resin and is found in the flowthrough. This can result from an inaccessible His-tag or overloading. While the flowthrough can be reapplied, optimizing the initial binding conditions is preferred.
• The refolded and eluted protein remains aggregation-prone. Precipitation can occur during the necessary dialysis or concentration. To mitigate this, maintain low temperatures, include stabilizing agents (e.g., glycerol, mild detergents), and avoid excessive concentration.
• This protocol prioritizes refolding and recovery over purity. The eluted protein is suitable for functional assays but typically requires further purification (e.g., gel filtration or ion-exchange chromatography) for structural or biophysical studies.
• The precise control of the dual gradient is essential and requires a chromatography system capable of programmable buffer mixing (e.g., ÄKTA Pure), which may not be available in all labs.
Troubleshooting
Problem 1: Low protein yield after refolding.
Possible causes: Insufficient solubilization of inclusion bodies or protein aggregation during refolding.
Solutions:
1) Increase chaotrope concentration in Buffer 3 or extend solubilization time.
2) Optimize the refolding gradient slope (use shallower gradients).
3) Add stabilizing additives (arginine 0.5 M, sucrose 0.4 M) to refolding buffers.
Problem 2: Poor binding to Ni-TED resin.
Possible causes: His-tag inaccessibility or incorrect buffer conditions.
Solutions:
1) Verify His-tag integrity by western blotting.
2) Ensure Buffer 3 contains 10–20 mM imidazole to reduce nonspecific binding.
3) Check pH stability during solubilization.
Problem 3: High nucleic acid contamination in the final preparation.
Possible causes: Incomplete benzonase treatment or insufficient washing.
Solutions:
1) Increase the benzonase incubation time to 45–60 min.
2) Add additional wash steps with Buffer 2 containing 1 mM MgCl2.
3) Include a DNA-specific precipitation step if necessary.
Problem 4: Inconsistent refolding between batches.
Possible causes: Variations in inclusion body purity or storage conditions.
Solutions:
1) Standardize the inclusion body preparation protocol across batches.
2) Use freshly prepared β-mercaptoethanol for each experiment.
3) Control and document storage time and conditions of inclusion bodies.
Supplementary information
The following supporting information can be downloaded here:
1. Dataset S1: Sequence
Acknowledgments
Author contributions: Conceptualization, A.V., A.K., M.E.; Investigation, A.V., D.P., Y.A., A.K., M.E.; Writing—Original Draft, A.V., D.P., Y.A.; Writing—Review & Editing, A.K., M.E.; Funding acquisition, M.P.
The work was carried out within the state assignment of NRC “Kurchatov Institute”.
Competing interests
The authors declare that they have no competing interests.
References
- Baneyx, F. (1999). Recombinant protein expression in Escherichia coli. Curr Opin Biotechnol. 10(5): 411–421. https://doi.org/10.1016/s0958-1669(99)00003-8
- Cabrita, L. D. and Bottomley, S. P. (2004). Protein expression and refolding – A practical guide to getting the most out of inclusion bodies. Biotechnol Annu Rev. 10: 31–50.https://doi.org/10.1016/s1387-2656(04)10002-1
- Georgiou, G. and Valax, P. (1996). Expression of correctly folded proteins in Escherichia coli. Curr Opin Biotechnol. 7(2): 190–197. https://doi.org/10.1016/s0958-1669(96)80012-7
- Nikitina, O. V., Agapova, Y. K., Petrenko, D. E. and Vlaskina, A. V. (2024). Features of the production of recombinant proteins containing disulfide bonds. Bulletin of the Military Innovative Technopolis "ERA". 5(2): 115–120. https://doi.org/10.56304/S2782375X24020189
- Vallejo, L. F. and Rinas, U. (2004). Strategies for the recovery of active proteins through refolding of bacterial inclusion body proteins. Microb Cell Fact. 3(1): 11. https://doi.org/10.1186/1475-2859-3-11
- Yamaguchi, H. and Miyazaki, M. (2014). Refolding Techniques for Recovering Biologically Active Recombinant Proteins from Inclusion Bodies. Biomolecules. 4(1): 235–251. https://doi.org/10.3390/biom4010235
- Katoh, S. and Katoh, Y. (2000). Continuous refolding of lysozyme with fed-batch addition of denatured protein solution. Process Biochem. 35(10): 1119–1124. https://doi.org/10.1016/s0032-9592(00)00145-x
- Jungbauer, A., Kaar, W. and Schlegl, R. (2004). Folding and refolding of proteins in chromatographic beds. Curr Opin Biotechnol. 15(5): 487–494. https://doi.org/10.1016/j.copbio.2004.08.009
- Gu, Z., Weidenhaupt, M., Ivanova, N., Pavlov, M., Xu, B., Su, Z. G. and Janson, J. C. (2002). Chromatographic Methods for the Isolation of, and Refolding of Proteins from, Escherichia coli Inclusion Bodies. Protein Expression Purif. 25(1): 174–179. https://doi.org/10.1006/prep.2002.1624
- Xi, H., Yuan, R., Chen, X., Gu, T., Cheng, Y., Li, Z., Jiang, C., Kong, W. and Wu, Y. (2016). Purification and on-column refolding of a single-chain antibody fragment against rabies virus glycoprotein expressed in Escherichia coli. Protein Expression Purif. 126: 26–32. https://doi.org/10.1016/j.pep.2016.05.004
- Kante, R. K., Vemula, S., Mallu, M. R. and Ronda, S. R. (2018). Efficient and easily scalable protein folding strong anion exchange chromatography for renaturation and simultaneous purification of recombinant human asparaginase from E. coli. Biotechnol Progr. 34(4): 1036–1044. https://doi.org/10.1002/btpr.2649
- Oganesyan, N., Kim, S.-H. and Kim, R. (2005). SOn-column Protein Refolding for Crystallization. J Struct Funct Genomics. 6(2–3): 177–182. https://doi.org/10.1007/s10969-005-2827-3
- Chang, P., Li, X., Lin, J., Li, C. and Li, S. (2022). scFv-oligopeptide chaperoning system-assisted on-column refolding and purification of human muscle creatine kinase from inclusion bodies. J Chromatogr B. 1209: 123410. https://doi.org/10.1016/j.jchromb.2022.123410
- Oyeleye, A. O., Mohd Yusoff, S. F., Abd Rahim, I. N., Leow, A. T. C., Saidi, N. B. and Normi, Y. M. (2020). Effective refolding of a cysteine rich glycoside hydrolase family 19 recombinant chitinase from Streptomyces griseus by reverse dilution and affinity chromatography. PLoS One. 15(10): e0241074. https://doi.org/10.1371/journal.pone.0241074
- De Bernardez Clark, E., Schwarz, E. and Rudolph, R. (1999). [15] Inhibition of aggregation side reactions during in vitro protein folding. Methods Enzymol. 309: 217–236. https://doi.org/10.1016/S0076-6879(99)09017-5
- Wu, W. Y., Mohanraju, P., Liao, C., Adiego-Pérez, B., Creutzburg, S. C., Makarova, K. S., Keessen, K., Lindeboom, T. A., Khan, T. S., Prinsen, S., et al. (2022). The miniature CRISPR-Cas12m effector binds DNA to block transcription. Mol Cell. 82(23): 4487–4502.e7. https://doi.org/10.1016/j.molcel.2022.11.003
- Omura, S. N., Nakagawa, R., Südfeld, C., Villegas Warren, R., Wu, W. Y., Hirano, H., Laffeber, C., Kusakizako, T., Kise, Y., Lebbink, J. H. G., et al. (2023). Mechanistic and evolutionary insights into a type V-M CRISPR–Cas effector enzyme. Nat Struct Mol Biol. 30(8): 1172–1182. https://doi.org/10.1038/s41594-023-01042-3
- Bigelyte, G., Duchovska, B., Zedaveinyte, R., Sasnauskas, G., Sinkunas, T., Dalgediene, I., Tamulaitiene, G., Silanskas, A., Kazlauskas, D., Valančauskas, L., et al. (2024). Innate programmable DNA binding by CRISPR-Cas12m effectors enable efficient base editing. Nucleic Acids Res. 52(6): 3234–3248. https://doi.org/10.1093/nar/gkae016
- Mohanraju, P., Oost, J., Jinek, M. and Swarts, D. (2018). Heterologous Expression and Purification of the CRISPR-Cas12a/Cpf1 Protein. Bio Protoc. 8(9): e2842. https://doi.org/10.21769/bioprotoc.2842
- Garner, M. M. and Revzin, A. (1981). A gel electrophoresis method for quantifying the binding of proteins to specific DNA regions: application to components of the Escherichia coli lactose operon regulatory system. Nucleic Acids Res. 9(13): 3047–3060. https://doi.org/10.1093/nar/9.13.3047
Article Information
Publication history
Received: Oct 19, 2025
Accepted: Dec 25, 2025
Available online: Jan 15, 2026
Published: Feb 5, 2026
Copyright
© 2026 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Vlaskina, A., Petrenko, D., Agapova, Y., Kuzminkova, A., Evteeva, M. and Patrushev, M. (2026). On-Column Dual-Gradient Refolding for Efficient Recovery of Insoluble Affinity-Tagged Recombinant Proteins. Bio-protocol 16(3): e5598. DOI: 10.21769/BioProtoc.5598.
Category
Biochemistry > Protein > Isolation and purification
Microbiology > Heterologous expression system > Escherichia coli
Biochemistry > Protein > Structure
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link







