- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Selective Enrichment and Identification of Cerebrospinal Fluid-Contacting Neurons In Vitro via PKD2L1 Promoter-Driven Lentiviral System
(*contributed equally to this work) Published: Vol 15, Iss 22, Nov 20, 2025 DOI: 10.21769/BioProtoc.5518 Views: 1343
Reviewed by: Oneil Girish BhalalaBaeksun KimAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Identification and Sorting of Adipose Inflammatory and Metabolically Activated Macrophages in Diet-Induced Obesity
Dan Wu [...] Weidong Wang
Oct 20, 2025 2243 Views

Optogenetic Approach for Investigating Descending Control of Nociception in Ex Vivo Spinal Cord Preparation
Volodymyr Krotov [...] Pavel Belan
Nov 5, 2025 1986 Views

Revisiting Primary Microglia Isolation Protocol: An Improved Method for Microglia Extraction
Jianwei Li [...] Guohui Lu
Dec 5, 2025 1475 Views
Abstract
Cerebrospinal fluid-contacting neurons (CSF-cNs) are a specialized group of multifunctional neurons located around the central canal of the spinal cord. They play critical roles in motor regulation, postural maintenance, and spinal cord injury repair. However, the molecular mechanisms underlying the multifunctionality of CSF-cNs remain poorly understood, partly due to the lack of established in vitro methods for their efficient selection and purification, which significantly hinders mechanistic investigations. In this study, we describe a standardized method using a PKD2L1 promoter-driven lentiviral system, which enables effective enrichment and identification of CSF-cNs in vitro through GFP labeling and puromycin selection. This protocol includes key steps such as construction of the PKD2L1 promoter-driven lentiviral vector, spinal cord tissue collection and digestion from neonatal mice, lentiviral infection, antibiotic selection, and immunofluorescence-based identification of CSF-cNs. Our method provides a reliable platform for obtaining high-purity CSF-cNs (>99%), which facilitates their functional and mechanistic studies for regenerative approaches in vitro.
Key features
• Enables specific labeling and selection of CSF-cNs using a constructed PKD2L1 promoter-driven lentiviral vector.
• Combines GFP-based fluorescence tracing and puromycin selection for efficient dual-mode enrichment of high-purity CSF-cNs.
• Provides a simple and reproducible workflow that facilitates in vitro isolation and validation of CSF-cNs for disease modeling and spinal cord injury repair.
Keywords: Cerebrospinal fluid-contacting neurons (CSF-cNs)Background
Cerebrospinal fluid-contacting neurons (CSF-cNs) are a population of multifunctional neurons located around the central canal of the spinal cord [1]. In addition to regulating motor behavior [2], maintaining posture [3], and contributing to central nervous system homeostasis [4], recent studies have revealed that CSF-cNs possess neural stem cell potential and may participate in tissue repair following spinal cord injury [5,6]. Nevertheless, the molecular mechanisms underlying their multifunctionality remain to be fully elucidated. The lack of standardized and specific in vitro methods for selective enrichment remains a major obstacle to comprehensive studies on the functional and regulatory features of these neurons.
Polycystic kidney disease 2-like 1 (PKD2L1), a calcium-permeable channel protein, is widely recognized as a specific molecular marker for CSF-cNs [1,7]. Its promoter has been successfully used in various model organisms to label and trace CSF-cNs in vivo [8–10]. In vitro, CSF-cNs can be obtained in transgenic mice through PKD2L1 promoter–driven fluorescent labeling combined with FACS sorting [11,12]. However, FACS-based methods often suffer from low post-sorting survival rates and limited expansion capacity [13,14], thereby restricting their generalizability and standardization. Therefore, it remains necessary to establish a standardized in vitro method for the selective enrichment and purification of CSF-cNs based on PKD2L1 expression.
In this study, we developed a lentiviral system driven by the mouse PKD2L1 promoter for efficient in vitro selection of CSF-cNs. This system incorporates dual reporter elements—green fluorescent protein (GFP) and puromycin resistance gene (PuroR)—enabling both fluorescence-based tracing and antibiotic-based selection. By infecting primary spinal cord cells with the virus, followed by puromycin selection and enrichment of GFP-positive cells, we obtained a high-purity CSF-cNs population. This system enables the acquisition of highly purified CSF-cNs under controlled conditions, thereby facilitating targeted interventions and pathway analyses, and providing a reliable platform for functional and mechanistic studies in vitro.
Materials and reagents
Biological materials
1. Neonatal C57BL/6 mice (postnatal day 1, Laboratory Animal Center, Guizhou Medical University)
2. PKD2L1-GFP-puromycin-luciferase lentivirus (lab-constructed)
3. psPAX2 (Addgene plasmid #12260; full sequence available at https://www.addgene.org/12260/)
4. pMD2.G (Addgene plasmid #12259; full sequence available at https://www.addgene.org/12259/)
5. HEK293T cells (ATCC, catalog number: CRL-11268; passage number 5 was used in this study)
Reagents
1. Papain (Sigma-Aldrich, catalog number: P3125); store at -20 °C
2. NeurobasalTM-A medium (Gibco, catalog number: 10888022); store at 4 °C
3. B-27TM supplement (Gibco, catalog number: 17504044); store at -20 °C
4. L-Glutamine (Gibco, catalog number: 25030081); store at -20 °C
5. Heparin, 0.2% solution (Sigma, catalog number: 07980); store at -20 °C
6. Penicillin-streptomycin solution (Gibco, catalog number: 15140122); store at 4 °C
7. Phosphate-buffered saline (PBS) (Gibco, catalog number: 10010023); store at room temperature
8. Accutase cell detachment solution (Innovative Cell Technologies, catalog number: AT104); store at 4 °C
9. Puromycin (Sigma-Aldrich, catalog number: P8833); store at -20 °C
10. Opti-MEMTM reduced serum medium (Gibco, catalog number: 31985070); store at 4 °C
11. Lipofectamine 3000 Transfection Reagent (Thermo Fisher Scientific, catalog number: L3000008; includes P3000 reagent); store at 4 °C
12. Poly-D-lysine (Sigma-Aldrich, catalog number: P7280); store at -20 °C
13. Paraformaldehyde, 4% solution (Sigma-Aldrich, catalog number: 158127); store at 4 °C
Growth factors
1. Recombinant human EGF (PeproTech, catalog number: AF-100-15); store at -20 °C, working concentration 20 ng/mL
2. Recombinant human bFGF (PeproTech, catalog number: AF-100-18B); store at -20 °C, working concentration 20 ng/mL
Immunostaining reagents
1. Primary antibody against PKD2L1 (Millipore, catalog number: AB9084); dilution 1:700, store at -20 °C
2. Anti-GFP primary antibody (mouse monoclonal) (Proteintech, catalog number: 66002-1-Ig); dilution 1:500, store at -20 °C
3. Goat anti-mouse IgG (H+L), Alexa FluorTM 488 secondary antibody (Thermo Fisher, catalog number: A11001); dilution 1:500, protect from light, store at -20 °C
4. Goat anti-rabbit IgG (H+L), Alexa FluorTM 594 secondary antibody (Thermo Fisher, catalog number: A11012); dilution 1:500, protect from light, store at -20 °C
5. DAPI staining solution (Solarbio, catalog number: C0065); store at -20 °C
6. Normal donkey serum (Yeasen Biotechnology, catalog number: 36136ES60), used at 5% in PBS for blocking, store at -20 °C, aliquoted
Solutions
1. Dissection medium (see Recipes)
2. Poly-D-lysine (PDL) Solution (see Recipes)
3. PBST washing buffer (see Recipes)
4. Nuclear antibody blocking buffer (for IF) (see Recipes)
5. Membrane antibody blocking buffer (for IF) (see Recipes)
6. DAPI staining solution (see Recipes)
7. Serum-free NSC medium (see Recipes)
Recipes
1. Dissection medium
a. Prepare by mixing DMEM high glucose with 1% L-Glutamine MAX and 5% penicillin-streptomycin.
b. Store at 4 °C for up to one month.
This solution is used for spinal cord tissue dissection and preservation prior to enzymatic digestion.
2. PDL solution
a. Dissolve 10 mg of poly-D-lysine powder in 10 mL of enzyme-free sterile water to obtain a 1 mg/mL stock solution.
b. Filter sterilize through a 0.22 μm membrane and aliquot.
c. Store at -20 °C for up to 6 months.
d. For use, dilute the stock solution tenfold to prepare a 0.1 mg/mL working solution. This working solution is used to coat culture plates and can be reused up to three times if stored properly at 4 °C for no longer than one week.
3. PBST washing buffer
a. Prepare 0.1% PBST by diluting Triton X-100 in PBS.
b. Store at 4 °C for up to one month.
This buffer is used for washing steps during immunofluorescence staining.
4. Nuclear antibody blocking buffer (for IF)
a. Prepare the blocking solution by mixing PBS with 0.5% Triton X-100, 5% BSA, and 22.5 mg/mL glycine.
b. Store at -20 °C for up to 6 months.
This is used to block nonspecific binding during nuclear-targeted immunofluorescence staining.
5. Membrane antibody blocking buffer (for IF)
a. Prepare the blocking solution using PBS supplemented with 0.3% Triton X-100, 5% BSA, and 22.5 mg/mL glycine.
b. Store at -20 °C for up to 6 months.
This is used for blocking in membrane-targeted immunofluorescence staining.
6. DAPI staining solution
a. Prepare DAPI solution by dissolving DAPI powder in PBS to a final concentration of 1 μg/mL.
b. Store at -20 °C for up to 6 months protected from light.
This solution is used for nuclear counterstaining by incubating cells for 5 min at room temperature in the dark.
7. Serum-free NSC medium
a. Mix the following components: Neurobasal-A medium, B27 supplement 2%, L-Glutamine 2 mM, penicillin-streptomycin solution 1%, recombinant human EGF 20 ng/mL, recombinant human bFGF 20 ng/mL, and heparin 0.2% solution.
b. The complete medium can be stored at 4 °C for up to 2 weeks, but freshly prepared medium is recommended to preserve growth factor activity.
Laboratory supplies
1. Ultra-low attachment 6-well plates (Corning, catalog number: 3471); used for suspension culture of CSF-cNs
2. Standard 24-well culture plates (Corning, catalog number: 3526); used for functional assays
3. Standard 96-well culture plates (Corning, catalog number: 3596); used for functional assays
Equipment
1. Class II A2 biosafety cabinet (ESCO)
2. CO2 incubator (Thermo Fisher Scientific, model: Heracell VIOS 160i)
3. Inverted fluorescence microscope (Olympus, model: IX73)
4. Confocal laser scanning microscope (Leica, model: SP8)
5. Benchtop centrifuge (Eppendorf, model: 5810R)
6. OptimaTM ultracentrifuge (Beckman Coulter, model: L-100K)
7. SW28 rotor (Beckman Coulter)
Software and datasets
1. UCSC Genome Browser (https://genome.ucsc.edu)
2. Ensembl Genome Browser (https://www.ensembl.org)
3. NCBI Database (https://www.ncbi.nlm.nih.gov)
4. BLAST (Basic Local Alignment Search Tool) (https://blast.ncbi.nlm.nih.gov/Blast.cgi)
5. ImageJ, NIH (https://imagej.nih.gov/ij/)
6. GraphPad Prism 10 (https://www.graphpad.com)
Procedure
The workflow comprises five main key modules: construction of a PKD2L1-promoter lentiviral vector, spinal cord tissue dissociation, lentiviral infection, antibiotic enrichment, and CSF-cNs identification (Figure 1).
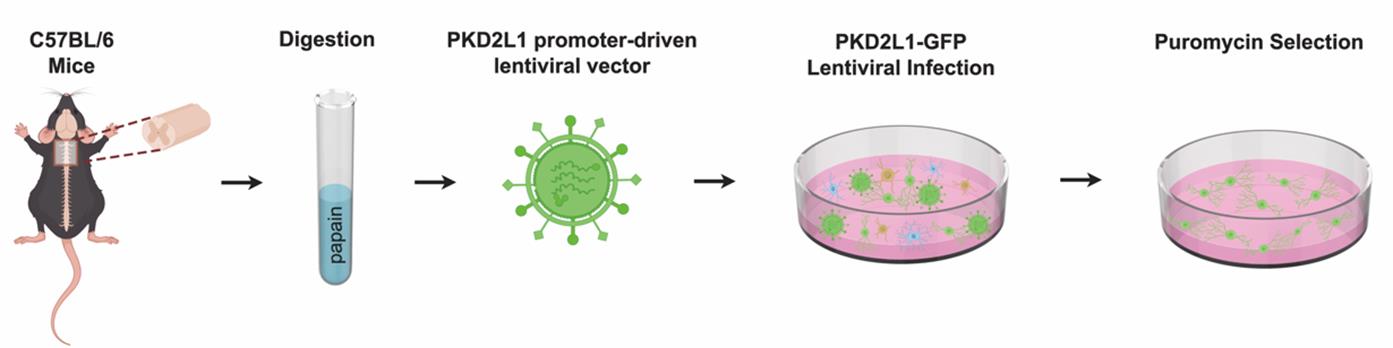
Figure 1. Workflow for in vitro selection of high-purity cerebrospinal fluid-contacting neurons (CSF-cNs) using PKD2L1 promoter-driven lentiviral vector. A stepwise schematic showing spinal cord tissue dissection, enzymatic digestion, PKD2L1-GFP lentiviral infection, and puromycin-based selection for reproducible enrichment of high-purity GFP+ CSF-cNs (purity >99%).
A. Construction of the PKD2L1-promoter lentiviral vector
1. Retrieve the genomic locus of mouse PKD2L1 (RefSeq: NM_181422) from UCSC, Ensembl, and NCBI databases.
2. Define the promoter region as a 1,026-bp fragment spanning -862 to +164 relative to Exon 1 (TSS = 0) and confirm its specificity in silico using NCBI BLAST/Primer-BLAST.
3. The 1,026-bp PKD2L1 promoter sequence from mouse is listed below:
AAGAGCTCTCGGACTCAGTTTATGCTGAAGCTTACAGTTCTAGTGCTAAGTCACGTCCCAGGAAAAGATTGACCAGATCATCTGGTACAGACAGATGTCACCTCACCCTTGCCCTAGGCCCCGAGTTATGCCTTCCCTACCCAGCCCCACATGCTCAAGAGGTCCATCTCTAAGGGTGTGTGTCACCATTAAGCCATGAAGCCAAACGTCAGGGAAAGAAAGCCAGCCTGGAACTCTGCATTCCTTTTCTTGTCACCAGCAGAGACCCTATCTCTTAAGATCTGACAGCCTGATTGCCTTCACCTGTGTCTTTCACTGACCTCTAAGGGAAGGGCACACTGGGAGCCTCCTTCTCCAGTCACCAGGGGACAGCTGAAGGAGGCACACTGAAGGGCCTGGGAGACTACTGATAGAGCTGTCAAGGAACACATCCCTGGGCTTGGTCCTCACCCTAATCCATGGTAGTCCCTTTAAGCCTTTTTTTTTTTTAAGTGGTATCAGCTCACACCCACCACAGAGCTCTGCAACCCTTTCTTCTCCTAGCTTCTCTACTGCTCCCTCCCTCCTCCCCCGTTGTCCCTGCCCTTTGACCAACTCAACAGTTGGAGATTTGAACATCCTACTCCGGGTTCTATCTGGCCCTAGCACACCCCCCTGCTCCCCTCCCCCATCCTTCTAGTCTGTCTGCATAACATCCTCACACCCCTTTCTTTTCTCAATTCCAGTTCCCCTTATAGTTCCATTCCTTTTGTGTCAGAGAGAGAGAGAGAGAGAGAGAGAGAGAGAGAGAGAGAGAGAAAAGAGAGAGAGAGAGCTCATTGAAGGATTGATGACCATCTGCATCCTTCCTGCTCCCTCTCCTATGTGAGCTCAGCTCCCCCACCCAGACAGGAGGCCGAGGTTGAAAGGATCAGCTGCTCTTAGACAATACTGCCTGGGCTCTCTGCTACCAGTGTCAGTCTGGGTCTTTGTCCCTGTGTCTCCTGTGAGAGTGGGCACCTGTGGTGGCAGGTTTCTACCTCCTGT
4. Insert the promoter upstream of a dual-reporter cassette—firefly luciferase (Fluc) followed by EGFP—in a lentiviral backbone (Figure 2).
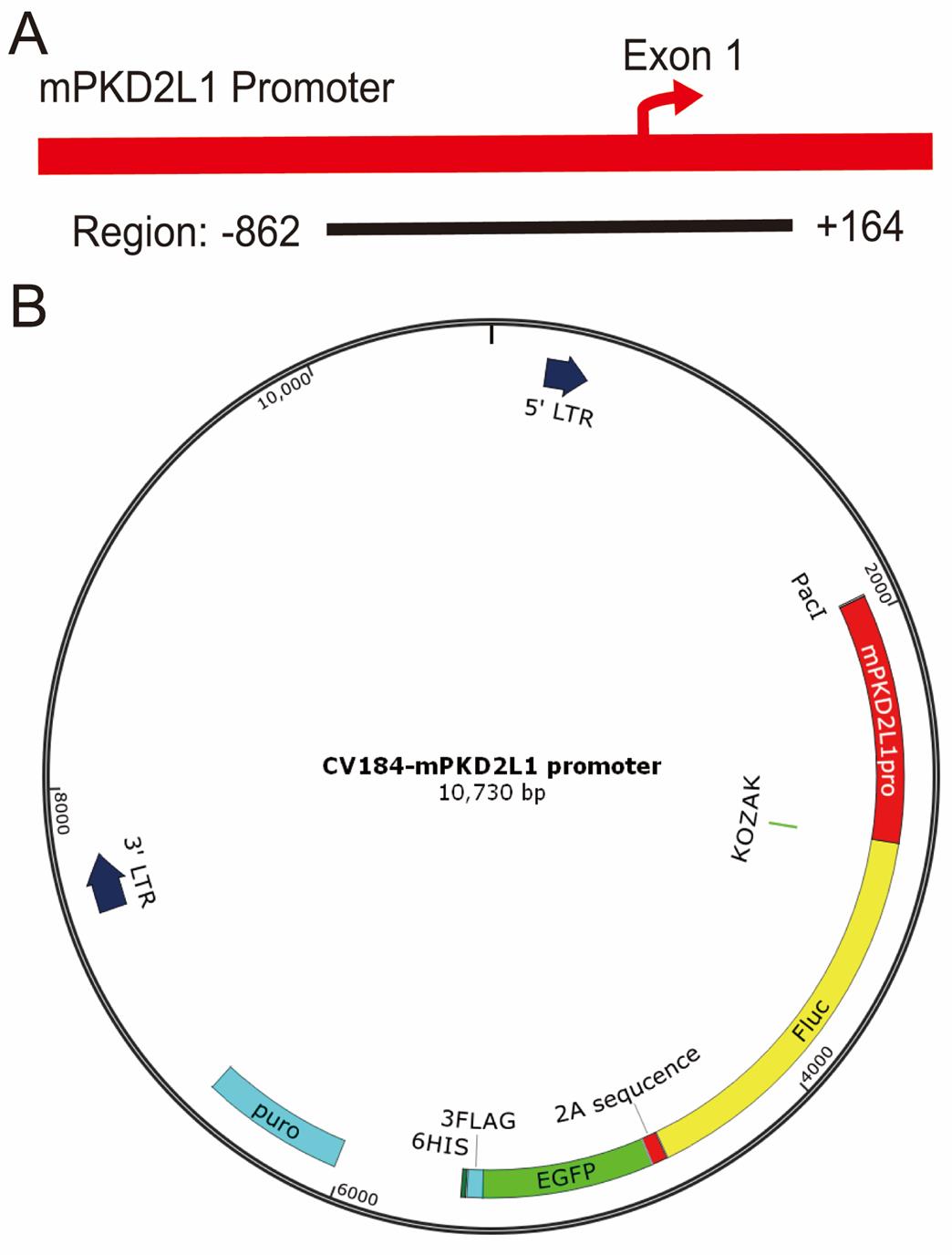
Figure 2. Schematic and structural diagrams of the PKD2L1 promoter-driven lentiviral vector. (A) Schematic representation of the Pkd2l1 promoter construct. The cloned fragment spans from the upstream promoter region (-862 bp) to +164 bp within the non-coding portion of Exon 1 (1,026 bp in total), encompassing the core promoter and proximal regulatory elements. The fragment does not contain any coding sequence (CDS). (B) Structural diagram of the PKD2L1 promoter lentiviral vector showing key elements including PuroR, GFP, and Fluc.
Note: A Fluc sequence is retained in the vector but is not used in this protocol; no in vivo imaging or luciferase assays are performed.
5. Verify the recombinant plasmid by Sanger sequencing to ensure that the promoter insert fragment and cloning junction sites are 100% identical to the reference sequence, with no mismatches allowed.
B. Lentivirus preparation
1. Co-transfect HEK293T cells using Lipofectamine 3000 with a three-plasmid system.
2. Prepare the transfection mix as follows:
a. Use 8 μg of total DNA in the following ratio: 4 μg of expression plasmid (PKD2L1 pro–GFP–PuroR), 3 μg of packaging plasmid (psPAX2), and 1 μg of envelope plasmid (pMD2.G).
b. Dilute the DNA with 16 μL of P3000 in 250 μL of Opti-MEM.
c. Dilute 12 μL of Lipofectamine 3000 in 250 μL of Opti-MEM.
d. Combine the DNA and Lipofectamine mixtures, incubate for 15 min at room temperature, and add dropwise to the cells.
3. Replace the medium 6 h post-transfection.
4. Collect the supernatant 48–72 h post-transfection, filter through a 0.45 μm membrane, and concentrate by ultracentrifugation:
a. Centrifuge at 100,000× g for 2 h at 4 °C using an SW28 rotor.
b. Discard the supernatant and resuspend the viral pellet in PBS.
c. Functionally titer the concentrated virus on HEK293T cells using 10-fold serial dilutions, followed by puromycin selection. Ensure the final titer is ≥1 × 108 TU/mL.
C. Quality control and titration of lentivirus
1. Verify the physical appearance of the viral solution: it should be a clear, light-pink solution with no viscosity.
2. Confirm sterility: infect HEK293T cells for 24 h. The culture should remain clear without microbial contamination.
3. Determine the functional titer using the puromycin-selection method:
a. Seed 4 × 104 HEK293T cells per well in a 96-well plate (100 μL).
b. Prepare 7–10 tenfold serial dilutions of the virus (see General notes for details) and add 90 μL per well.
c. After 72 h, add 5 μg/mL puromycin and incubate for 24 h.
d. Count surviving cells and calculate the titer.
D. Spinal cord tissue collection
1. Use neonatal (postnatal day 1) C57BL/6 mice.
2. Perform hypothermic anesthesia on ice, sterilize with 75% ethanol, and decapitate.
3. Dissect the spinal cord on ice [15,16] and transfer to dissection medium (DMEM-HG supplemented with 1% L-Glutamine MAX and 5% penicillin-streptomycin).
4. Mince the tissue into ~0.5 mm3 fragments.
E. Enzymatic dissociation and cell resuspension
1. Digest with papain at 37 °C for 30 min.
2. Centrifuge at 200× g for 5 min and discard the supernatant.
3. Resuspend the pellet in serum-free NSC medium (see Recipes).
F. Cell culture and lentiviral infection
1. Seed the dissociated spinal cord cells into ultra-low attachment 6-well plates and maintain them in culture under suspension conditions.
2. Plate the cells onto 24-well plates pre-coated with 0.1 mg/mL poly-D-lysine, which promotes cell adhesion.
3. Infect with lentivirus at a multiplicity of infection (MOI) of 20 (see General notes for details) for 12–24 h.
4. Replace with fresh medium; after 3 days, add puromycin (1–2 μg/mL) and select for 48–72 h to obtain GFP-positive cells (Figure 3A).
G. Identification of CSF-cNs
1. After puromycin selection (1–2 μg/mL for 48–72 h), monitor GFP expression under a fluorescence microscope. A marked increase in GFP+ cells indicates successful enrichment (Figure 3B).
2. Perform immunofluorescence staining using an anti-PKD2L1 antibody to confirm CSF-cNs identity.
a. Fix cells with 4% paraformaldehyde for 15 min at room temperature.
b. Block with 5% donkey serum in PBS for 30 min.
c. Incubate with primary antibodies against PKD2L1 (1:300) and GFP (1:500) overnight at 4 °C. Because fixation attenuates endogenous GFP signals after viral transduction, an anti-GFP antibody is included for signal amplification.
d. Wash the cells and incubate with secondary antibodies for 1 h at room temperature, including Alexa Fluor 594–conjugated secondary antibody for PKD2L1 detection and Alexa Fluor 488–conjugated secondary antibody for GFP detection.
e. Counterstain nuclei with DAPI (1 μg/mL) for 5 min.
f. Visualize under a fluorescence microscope. Colocalization of PKD2L1 (red) and GFP (green) confirms the identity of CSF-cNs (Figure 3C).
g. Quantify the percentage of PKD2L1+ cells among GFP+ cells to evaluate labeling specificity (Figure 3D).
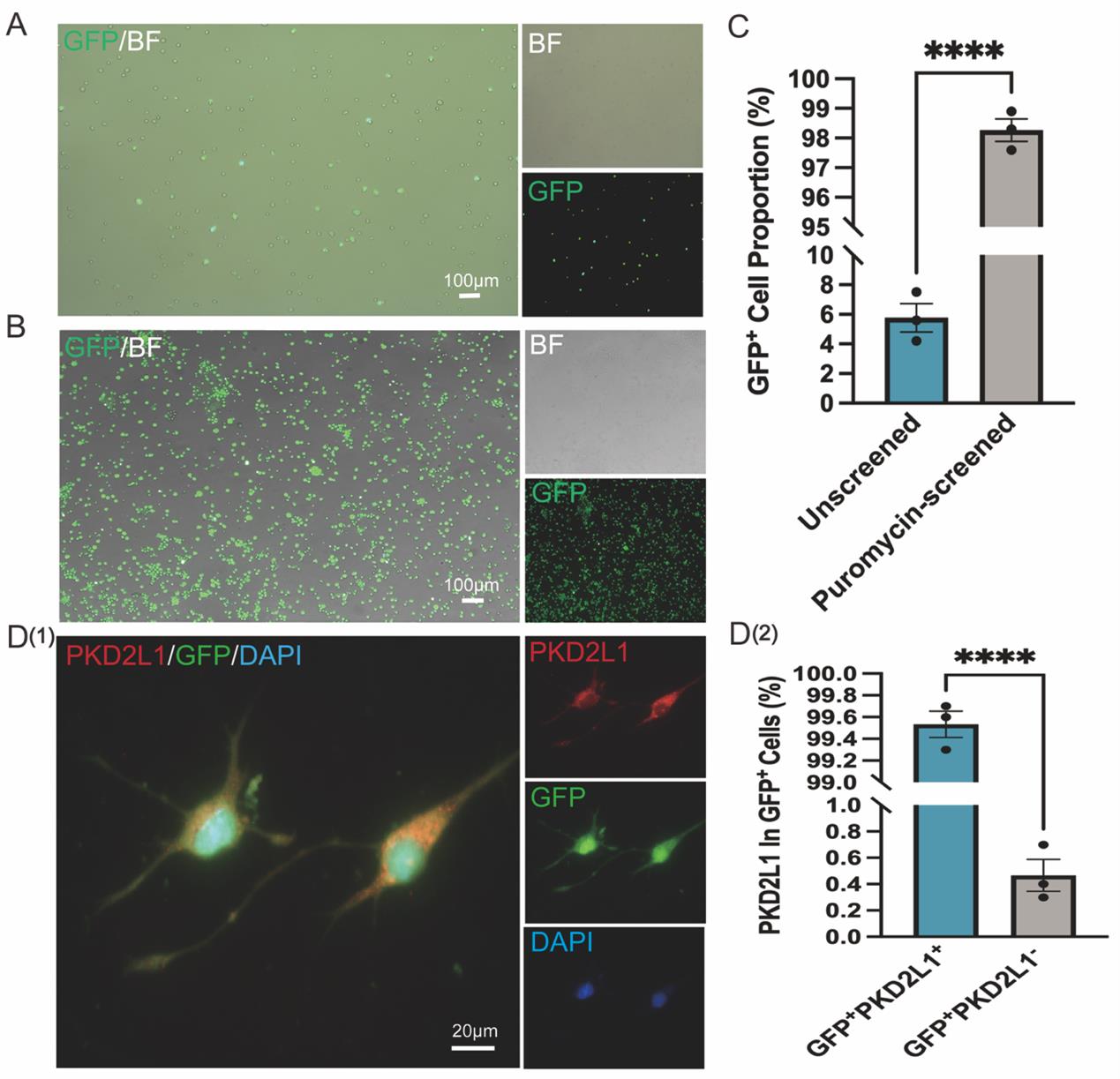
Figure 3. Purification and identification of cerebrospinal fluid-contacting neurons (CSF-cNs) using PKD2L1 promoter-driven lentiviral vector enriched by (GFP+ + puromycin resistance) dual-mode selection strategy. (A) Representative images showing sparse GFP+ CSF-cNs following lentiviral infection without puromycin selection. Left: merged GFP and brightfield (BF) image; top right: BF image; bottom right: GFP fluorescence. Scale bar: 100 μm. (B) Representative images showing enriched GFP+ CSF-cNs after puromycin selection. Left: merged GFP and BF image; top right: BF image; bottom right: GFP fluorescence. Scale bar: 100 μm. (C) Quantification of GFP+ cell percentage before (unscreened) and after puromycin selection (puromycin-selected), demonstrating a significant increase in purity from 5.7% to 98.6% (n = 3, mean ± SEM). Statistical analysis was performed using an unpaired two-tailed Student’s t-test. ****P < 0.0001. (D-1) Representative immunofluorescence images of puromycin-selected CSF-cNs showing co-expression of PKD2L1 (red) and GFP (green), with DAPI-stained nuclei (blue). Right panels display individual fluorescence channels. Scale bar: 20 μm. (D-2) Quantification shows that PKD2L1+ cells account for more than 99% of GFP+ CSF-cNs (n = 3). Data are presented as mean ± SEM. Statistical analysis was performed using an unpaired two-tailed Student’s t-test. ****P < 0.0001.
Validation of protocol
This protocol has been successfully and reproducibly applied to spinal cord tissue samples from five independent neonatal mice, consistently yielding GFP+ PKD2L1+ CSF-cNs. Across three independent experiments, both infection efficiency and selection purity were consistently high. Following puromycin selection, the proportion of GFP+ cells increased from an average of 5.8% to over 98% (Figure 3C), and more than 99% of the GFP+ cells co-expressed PKD2L1 (Figure 3D), indicating efficient and specific enrichment of CSF-cNs. This method was also applied in our previously published study investigating CSF-cNs transplantation for spinal cord injury repair, demonstrating strong reproducibility and broad applicability [5].
Data analysis
Quantification of immunofluorescence images was performed using ImageJ (NIH). For each group, three randomly selected fields were analyzed to determine the percentage of GFP+/PKD2L1+ double-positive cells among the total GFP+ population. Samples were collected from five independent neonatal mice as biological replicates, with at least three fields per mouse serving as technical replicates. Data are presented as mean ± standard error of the mean (SEM). Graphs were generated using GraphPad Prism 10. Statistical analyses were performed using an unpaired two-tailed Student’s t-test, and P < 0.05 was considered statistically significant. The results showed that the purity of selected cells consistently exceeded 98%, with minimal intergroup variability, indicating strong reproducibility and consistency of the protocol.
General notes and troubleshooting
General notes
1. Promoter fragment selection: We selected the -862/+164 fragment (1,026 bp in total) as the PKD2L1 promoter, following the common strategy of including approximately 1 kb of upstream promoter sequence and a short overlap with the non-coding portion of Exon 1 to cover the core promoter and proximal regulatory elements. Its specificity was further verified by BLAST against the mouse genome.
2. Serial dilutions for viral titration: To prepare 7–10 tenfold serial dilutions, transfer 100 μL of viral stock into 900 μL of Opti-MEM medium in a 1.5 mL EP tube to obtain the 10-1 dilution. Mix thoroughly, then take 100 μL from this tube and add it to a new tube containing 900 μL of Opti-MEM to obtain the 10-2 dilution. Repeat sequentially until the desired dilution (up to 10-8) is reached.
3. Viral titer calculation: When performing viral titration, the following example can be used for clarity: If three viable cells are observed in the 10-5 μL dilution, titer = 3/10-5 μL = 3 × 105 TU/µL = 3 × 108 TU/mL. This illustrates how to estimate the functional viral titer from serial dilution results.
4. Optimization and validation of MOI = 20: In pilot experiments, CSF-cNs were seeded at a density of 6,000 cells per well in 96-well plates and infected with lentivirus at MOI values ranging from 0 to 200. The results showed that MOI = 20 consistently achieved >90% GFP+ cells with good viability, whereas higher MOI values markedly reduced survival. Therefore, MOI = 20 was established as the standard infection condition in this study. To ensure reproducibility, viral titers were determined by a puromycin resistance–based assay (≥1 × 108 TU/mL), and the required viral volume was calculated using the formula MOI = (TU × volume)/cell number to guarantee that the infection condition was maintained at MOI = 20.
5. Lentiviral titer and infection time: A lentiviral titer of ≥1 × 108 TU/mL is recommended. To ensure sufficient expression of GFP driven by the PKD2L1 promoter, an extended infection duration of up to 48 h may be required.
6. Puromycin concentration optimization: The optimal puromycin concentration may vary depending on cell density and viral infection efficiency. It is recommended to perform a pilot titration test. A working range of 1–2 μg/mL is generally effective while minimizing cytotoxicity.
7. Luciferase element: The luciferase (Luc) sequence was retained in the lentiviral construct for potential future applications (e.g., in vivo bioluminescence imaging and dynamic cell monitoring after transplantation). However, Luc was not used in the present protocol, and its presence does not affect the functions of GFP and puromycin in CSF-cNs enrichment and identification.
8. Spinal cord collection: In this protocol, the entire spinal cord was collected without precise distinction of the cervical–thoracic or cervical–medullary junctions, as this does not affect the isolation of CSF-cNs.
9. Control considerations: This study did not include a systematic analysis of the proportion of PKD2L1+ cells within GFP-positive and GFP-negative populations prior to puromycin selection. Such data would serve as important negative controls, and future studies should incorporate this analysis to further strengthen protocol validation.
10. Promoter fragment features: The cloned PKD2L1 promoter fragment (-862/+164 bp) extends 164 bp into Exon 1 but does not contain any coding sequence (CDS); thus, it will not produce an Exon1::GFP fusion protein. Nevertheless, to further ensure labeling specificity, future studies should include validation using additional established CSF-cNs markers (such as GATA3 or GABA).
Troubleshooting
| Problem | Possible cause(s) | Solution(s) |
|---|---|---|
| Low or no GFP expression in CSF-cNs. | Lentiviral titer is too low, or infection duration is insufficient. | Increase viral concentration and/or extend infection duration up to 48 h. |
| High cell death during puromycin selection of CSF-cNs. | Puromycin concentration is too high. | Perform a puromycin kill curve in advance; reduce concentration to within 1–2 μg/mL. |
| Low correlation between GFP expression and PKD2L1 immunostaining in CSF-cNs. | GFP signal may not directly reflect PKD2L1 protein expression. | Validate sorted GFP+ cells by immunostaining with anti-PKD2L1 antibody. |
Note: “Puromycin kill curve” refers to a preliminary experiment used to determine the minimum concentration of puromycin required to completely eliminate untransfected control cells within 3–5 days. This concentration is then applied in subsequent selection experiments.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant No. 82160249) and the NSFC Cultivation Project of the Affiliated Hospital of Guizhou Medical University (Grant No. gyfynsfc-2022-52). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Competing interests
The authors declare no competing interests.
Ethical considerations
All animal procedures were conducted in accordance with institutional and national guidelines for the care and use of laboratory animals. The protocol was approved by the Animal Ethics Committee of Guizhou Medical University.
References
- Wyart, C., Carbo-Tano, M., Cantaut-Belarif, Y., Orts-Del’Immagine, A. and Böhm, U. L. (2023). Cerebrospinal fluid-contacting neurons: multimodal cells with diverse roles in the CNS. Nat Rev Neurosci. 24(9): 540–556. https://doi.org/10.1038/s41583-023-00723-8
- Wu, M. Y., Carbo-Tano, M., Mirat, O., Lejeune, F. X., Roussel, J., Quan, F. B., Fidelin, K. and Wyart, C. (2021). Spinal sensory neurons project onto the hindbrain to stabilize posture and enhance locomotor speed. Curr Biol. 31(15): 3315–3329.e5. https://doi.org/10.1016/j.cub.2021.05.042
- Sternberg, J. R., Prendergast, A. E., Brosse, L., Cantaut-Belarif, Y., Thouvenin, O., Orts-Del’Immagine, A., Castillo, L., Djenoune, L., Kurisu, S., McDearmid, J. R., et al. (2018). Pkd2l1 is required for mechanoception in cerebrospinal fluid-contacting neurons and maintenance of spine curvature. Nat Commun. 9(1): e1038/s41467–018–06225–x. https://doi.org/10.1038/s41467-018-06225-x
- Prendergast, A. E., Jim, K. K., Marnas, H., Desban, L., Quan, F. B., Djenoune, L., Laghi, V., Hocquemiller, A., Lunsford, E. T., Roussel, J., et al. (2023). CSF-contacting neurons respond to Streptococcus pneumoniae and promote host survival during central nervous system infection. Curr Biol. 33(5): 940–956.e10. https://doi.org/10.1016/j.cub.2023.01.039
- Luo, Z., Shangguan, Z., Cao, L., Zhang, Y., Li, Q., Shi, X., Fu, J., Wang, C., Dou, X., Tan, W., et al. (2025). Cerebrospinal fluid-contacting neurons: a promising source for adult neural stem cell transplantation in spinal cord injury treatment. Front Cell Dev Biol. 13: e1549194. https://doi.org/10.3389/fcell.2025.1549194
- Cao, L., Huang, M. Z., Zhang, Q., Luo, Z. R., Zhang, Y., An, P. J., Yang, L. L., Tan, W., Wang, C. Q., Dou, X. W., et al. (2022). The neural stem cell properties of Pkd2l1+ cerebrospinal fluid-contacting neurons in vivo. Front Cell Neurosci. 16: e992520. https://doi.org/10.3389/fncel.2022.992520
- Petracca, Y. L., Sartoretti, M. M., Di Bella, D. J., Marin-Burgin, A., Carcagno, A. L., Schinder, A. F. and Lanuza, G. M. (2016). The late and dual origin of cerebrospinal fluid-contacting neurons in the mouse spinal cord. Development. 143(5): e129254. https://doi.org/10.1242/dev.129254
- Böhm, U. L., Prendergast, A., Djenoune, L., Nunes Figueiredo, S., Gomez, J., Stokes, C., Kaiser, S., Suster, M., Kawakami, K., Charpentier, M., et al. (2016). CSF-contacting neurons regulate locomotion by relaying mechanical stimuli to spinal circuits. Nat Commun. 7(1): e1038/ncomms10866. https://doi.org/10.1038/ncomms10866
- Nakamura, Y., Kurabe, M., Matsumoto, M., Sato, T., Miyashita, S., Hoshina, K., Kamiya, Y., Tainaka, K., Matsuzawa, H., Ohno, N., et al. (2023). Cerebrospinal fluid-contacting neuron tracing reveals structural and functional connectivity for locomotion in the mouse spinal cord. eLife. 12: e83108. https://doi.org/10.7554/elife.83108
- Yue, W. W. S., Touhara, K. K., Toma, K., Duan, X. and Julius, D. (2024). Endogenous opioid signalling regulates spinal ependymal cell proliferation. Nature. 634(8033): 407–414. https://doi.org/10.1038/s41586-024-07889-w
- Wang, S., He, Y., Zhang, H., Chen, L., Cao, L., Yang, L., Wang, C., Pan, Y., Tang, Q., Tan, W., et al. (2021). The Neural Stem Cell Properties of PKD2L1+ Cerebrospinal Fluid-Contacting Neurons in vitro. Front Cell Neurosci. 15: e630882. https://doi.org/10.3389/fncel.2021.630882
- Cao, L., Zhang, H. Q., He, Y. Q., An, P. J., Yang, L. L., Tan, W., Liu, G., Wang, C. Q., Dou, X. W., Li, Q., et al. (2023). Culture of cerebrospinal fluid-contacting neurons from neonatal mouse spinal cord. Cell Tissue Banking. 25(2): 443–452. https://doi.org/10.1007/s10561-023-10098-w
- Janssens, S., Schotsaert, M., Manganaro, L., Dejosez, M., Simon, V., García‐Sastre, A. and Zwaka, T. P. (2018). FACS‐Mediated Isolation of Neuronal Cell Populations From Virus‐Infected Human Embryonic Stem Cell–Derived Cerebral Organoid Cultures. Curr Protoc Stem Cell Biol. 48(1): e65. https://doi.org/10.1002/cpsc.65
- Naama, M., Rahamim, M., Zayat, V., Sebban, S., Radwan, A., Orzech, D., Lasry, R., Ifrah, A., Jaber, M., Sabag, O., et al. (2023). Pluripotency-independent induction of human trophoblast stem cells from fibroblasts. Nat Commun. 14(1): e1038/s41467–023–39104–1. https://doi.org/10.1038/s41467-023-39104-1
- Eldeiry, M., Yamanaka, K., Reece, T. B. and Aftab, M. (2017). Spinal Cord Neurons Isolation and Culture from Neonatal Mice. J Visualized Exp. 125: e3791/55856. https://doi.org/10.3791/55856
- Anderson, K. N., Potter, A. C., Piccenna, L. G., Quah, A. K., Davies, K. E. and Cheema, S. S. (2004). Isolation and culture of motor neurons from the newborn mouse spinal cord. Brain Res Protoc. 12(3): 132–136. https://doi.org/10.1016/j.brainresprot.2003.10.001
Article Information
Publication history
Received: Jul 30, 2025
Accepted: Oct 12, 2025
Available online: Oct 31, 2025
Published: Nov 20, 2025
Copyright
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Tan, W., Shangguan, Z., Jing, S., Shi, X., Li, Q., Li, H., Zhang, H., Luo, Z., Wang, C., Dou, X. and Li, Q. (2025). Selective Enrichment and Identification of Cerebrospinal Fluid-Contacting Neurons In Vitro via PKD2L1 Promoter-Driven Lentiviral System. Bio-protocol 15(22): e5518. DOI: 10.21769/BioProtoc.5518.
Category
Neuroscience > Sensory and motor systems > Spinal cord
Cell Biology > Cell isolation and culture > Cell isolation
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








