- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Use of a High-Affinity Ubiquitin-Binding Domain to Detect and Purify Ubiquitinated Substrates and Their Interacting Proteins
(*contributed equally to this work) Published: Vol 15, Iss 17, Sep 5, 2025 DOI: 10.21769/BioProtoc.5426 Views: 3542
Reviewed by: Kazem NouriAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
Detection of Hog1 Phosphorylation in Candida albicans in Response to an Antifungal Protein
Brigitte ME Hayes and Nicole L van der Weerden
Sep 20, 2014 9240 Views
Far-western Blotting Detection of the Binding of Insulin Receptor Substrate to the Insulin Receptor
Jinghua Peng [...] Ling He
Feb 20, 2023 2309 Views

Monitoring Endocytosis of Integral Membrane Proteins Using Western Blot-Based Detection of Biotinylated Antibody Uptake
Alexandra Graninger and Prasanna Satpute-Krishnan
Nov 20, 2025 1742 Views
Abstract
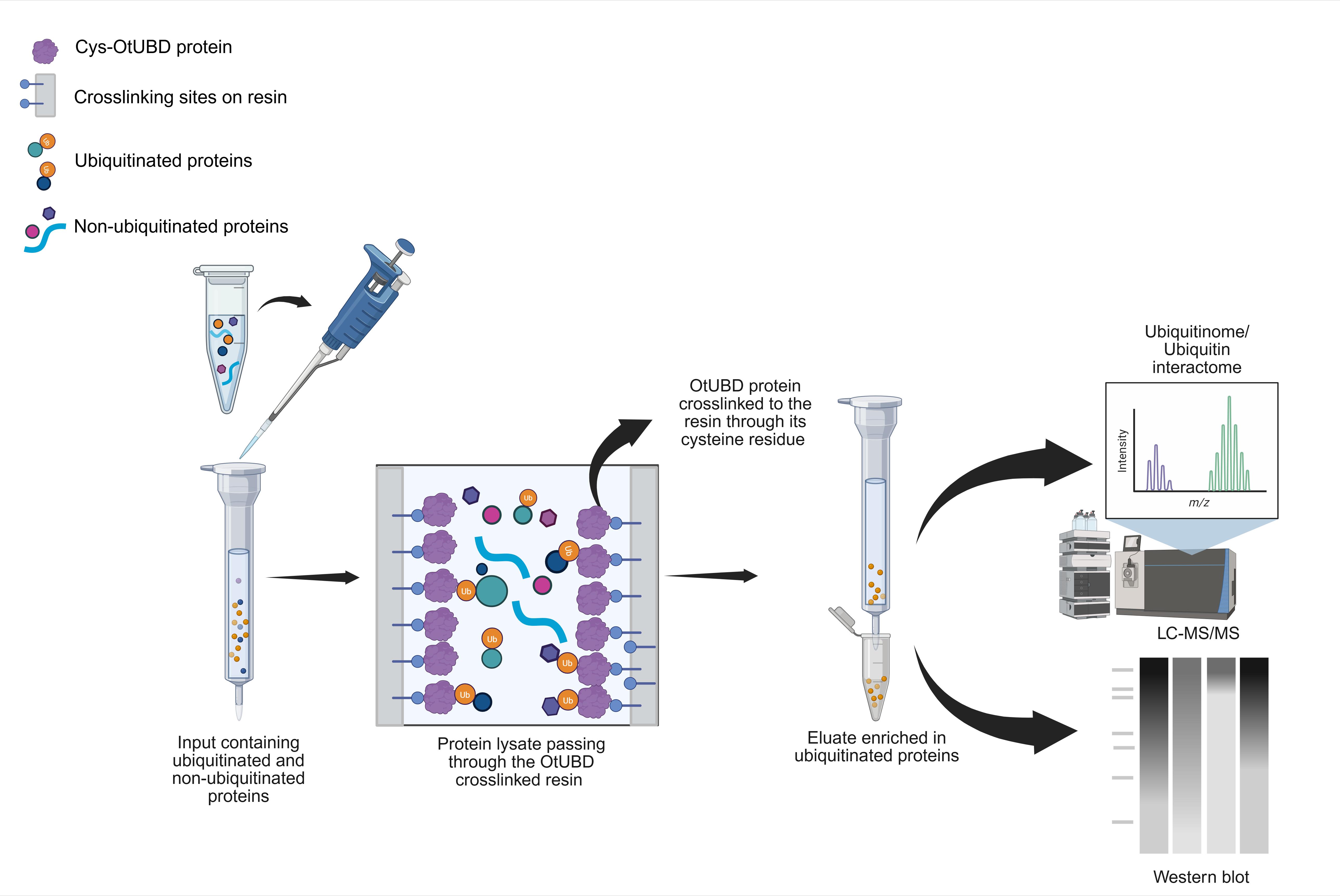
OtUBD is a high-affinity ubiquitin-binding domain (UBD) derived from a large protein produced by the microorganism Orientia tsutsugamushi. The following protocol describes a step-by-step process for the enrichment of ubiquitinated proteins from baker's yeast and mammalian cell lysates using OtUBD. The OtUBD affinity resin can strongly enrich both mono- and poly-ubiquitinated proteins from crude lysates. The protocol further describes the use of different buffer formulations to specifically enrich for proteins covalently modified by ubiquitin with or without proteins that associate with them. Combining different OtUBD-mediated enrichment protocols with liquid chromatography–tandem mass spectrometry (LC–MS/MS) helps distinguish the pool of covalently ubiquitinated proteins (the ubiquitinome) from ubiquitin- or ubiquitinated protein-interacting proteins (the ubiquitin interactome). The OtUBD tool described in the protocol has been used successfully with downstream applications such as immunoblotting and differential proteomics. It provides researchers with a versatile and economical tool for the study of ubiquitin biology.
Key features
• The protocol offers a native workflow and a denaturing workflow for enrichment of ubiquitinated proteins with or without noncovalently associated proteins, respectively.
• Included in the protocol are different resin compositions, lysate preparation methods, elution methods, and pulldown formats to suit different experimental needs.
• The protocol has been used in various applications, including immunoblotting, proteomics, and UbiCREST (ubiquitin chain restriction), and works with all types of ubiquitin conjugates.
• The protocol was developed and tested with budding yeast and mammalian cell lysates but can be adapted to other biological samples and organisms.
Keywords: UbiquitinGraphical overview
Background
The small protein ubiquitin exists in all eukaryotic organisms and is an essential post-translational protein modifier. Substrate proteins are covalently attached to ubiquitin via the coordinated reactions of three enzymes: the E1 enzyme, which mediates ATP-dependent activation of the ubiquitin C-terminus [1], the E2 conjugating enzyme, which carries the activated ubiquitin via a thioester bond [2], and finally, the E3 ligase [3], which directly interacts with the substrate and enables the transfer of ubiquitin to the substrate, which is usually, but not always, a protein [4]. Ubiquitin itself can be conjugated to other ubiquitin molecules via their N-terminal amino group or lysine side chains, forming distinct polymers that can specify interactions with different ubiquitin-binding domains [5]. For example, proteins modified by K48-linked polyubiquitin chains are typically recognized by the proteasome, where they are degraded [6], while many proteins marked with K63-linked polyubiquitin chains function in the endocytic or DNA repair pathways [7].
The diversity in ubiquitin chain topologies and linkages, as well as the attachment of single ubiquitin molecules, allows a corresponding diversity in substrate protein fates in many cellular pathways. Moreover, defective ubiquitination has been associated with many diseases, including neurodegenerative disorders and various cancers [8,9]. Thus, identifying ubiquitinated proteins and ways in which their modification might change under different conditions is often a key step in understanding cellular regulatory mechanisms and maladies resulting from their dysregulation.
At this point, many methods of enriching ubiquitinated proteins from lysates have been developed [10–12]. These each have specific advantages and disadvantages. For example, ectopic (over)expression of epitope-tagged ubiquitin, which is widely used for affinity purification of ubiquitinated targets using anti-epitope antibodies, may lead to spurious protein ubiquitination patterns, while the use of antibodies to ubiquitin for purification of conjugates at endogenous levels may lack sufficient sensitivity or specificity [13]. Tandem ubiquitin-binding entities (TUBEs), which link multiple low-affinity UBDs in a single polypeptide, are highly efficient in purifying polymeric ubiquitin, but work poorly against monoubiquitinated proteins, which usually constitute a large fraction of ubiquitinated proteins in mammalian cells and tissues [14,15]. Antibodies raised against remnant peptides derived from trypsin-digested ubiquitin conjugated to lysine side chains of substrate proteins have been extremely effective in the proteomic identification of ubiquitination sites in proteins, but the antibodies are expensive and only reveal lysine modifications and not modification of other protein side-chain sites (such as serine or threonine) or non-protein substrates [4,16].
Earlier research from our group revealed that a UBD (OtUBD) found in a large deubiquitinase protein (OtDUB) from the bacterial pathogen Orientia tsutsugamushi exhibits very high affinity for ubiquitin, with a dissociation constant in the low nanomolar range [17]. This prompted the development of an OtUBD affinity resin for the enrichment of ubiquitinated proteins from complex biological samples [18]. In the present protocol, we describe step-by-step procedures for purifying the recombinant OtUBD polypeptide and for the preparation of the OtUBD resin. We detail methods to enrich ubiquitinated proteins from either baker’s yeast or human tissue culture cells, including the use of strong denaturants to distinguish directly ubiquitinated proteins from proteins interacting noncovalently with ubiquitin or ubiquitin-modified proteins.
Materials and reagents
Plasmids used to purify OtUBD for the creation of affinity resin
1. pRT498-OtUBD (Addgene, plasmid #190089)
2. pET21a-cys-His6-OtUBD (Addgene, plasmid #190091)
Reagents
1. BactoTM peptone (Gibco, catalog number: 211677)
2. BactoTM yeast extract (Gibco, catalog number: 212750)
3. BactoTM tryptone (Gibco, catalog number: 211705)
4. Bromophenol blue (Sigma-Aldrich, catalog number: B0126)
5. Agar (Difco, catalog number: 214010)
6. D-glucose anhydrous (Fischer chemicals, catalog number: D16-10)
7. Ampicillin sodium salt (Sigma-Aldrich, catalog number: A0166)
8. Kanamycin sulfate from Streptomyces kanamyceticus (Sigma-Aldrich, catalog number: K1637)
9. Phosphate-buffered saline (DPBS), 10×, Dulbecco's formula (Thermo Scientific, catalog number: J61917.AP)
10. Dithiothreitol (DTT) (Ultrapure AmericanBio, catalog number: AB00490)
11. DNase I (Roche, catalog number: 10104159001)
12. cOmplete EDTA-free protease inhibitor cocktail (Roche, catalog number: 11873580001)
13. Ethylenediaminetetraacetic acid disodium salt dihydrate (EDTA) (Sigma-Aldrich, catalog number: E5134)
14. Glycine (Fisher Bioreagents, catalog number: BP381-5)
15. Glycerol (Sigma-Aldrich, catalog number: G7893-4L)
16. Hydrochloric acid (HCl) 36.5%–38% (J.T. Baker, catalog number: 9535-33)
17. Imidazole (Sigma-Aldrich, catalog number: I202)
18. Isopropyl β-D-1-thiogalactopyranoside (IPTG) (RPI, catalog number: 156000)
19. L-cysteine (Sigma-Aldrich, catalog number: C7352)
20. Lysozyme (RPI, catalog number: L38100)
21. N-ethylmaleimide (NEM) (Sigma-Aldrich, catalog number: E3876)
22. Ni-NTA agarose (Qiagen, catalog number: 30230)
23. Trizma base (Tris base) for Tris buffers (Sigma-Aldrich, catalog number: T6066)
24. Tween 20 (Sigma-Aldrich, catalog number: P7949)
25. Triton-X 100 (AmericanBio, catalog number: AB02025)
26. Sodium azide (Sigma-Aldrich, catalog number: S2002)
27. SulfoLinkTM coupling resin (Thermo Scientific, catalog number: 20402)
28. Sodium chloride (NaCl) (Sigma-Aldrich, catalog number: 746398)
29. Sodium dodecyl sulphate (SDS) (GFS Chemicals, catalog number: 2288)
30. Phenylmethylsulfonyl fluoride (PMSF) (MP Biomedicals, catalog number: 195381)
31. Tris(2-carboxyethyl) phosphine hydrochloride (TCEP) (Sigma-Aldrich, catalog number: C4706)
32. Glass beads, acid washed (Sigma-Aldrich, catalog number: G8772)
33. Sodium hydroxide (NaOH) (Macron, catalog number: 7708-06)
34. GelCode Blue (Thermo Scientific, catalog number: 24594)
35. Urea (Sigma-Aldrich, catalog number: U5378)
36. PierceTM Bradford Protein Assay kit (Thermo Scientific, catalog number: 23200)
37. PierceTM BCA Protein Assay kit (Thermo Scientific, catalog number: 23225)
38. Nonfat milk (Chem Cruz, catalog number: SC-2325)
39. Immobilon-P PVDF membrane (Millipore, catalog number: IPVH00010)
40. Primary antibodies used in this study: rabbit polyclonal anti-ubiquitin antibody (Dako, Denmark, discontinued, 1:2,000 dilution), monoclonal mouse anti-ubiquitin antibody P4D1 (Enzo, USA, 1:1,000 or Invitrogen, USA, 1:4,000), and rabbit anti-ubiquitin antibody (E412J) (Cell Signalling, USA, 1:4,000)
41. For detection of rabbit primary antibodies, the HRP-linked anti-rabbit IgG secondary antibody (GE Healthcare, catalog number: NA934) was used at a dilution of 1:5,000 or 1:10,000
42. For detection of mouse primary antibodies, the HRP-linked anti-mouse secondary antibody (GE Healthcare, catalog number: NXA931V) was used at a dilution of 1:10,000
43. His-tagged tobacco etch virus (TEV) protease (purified in-house as a 2 mg/mL stock or NEB, catalog number: P8112)
44. SYPRO Ruby protein gel stain (Invitrogen, catalog number: S12000)
45. 4%–15% Mini-PROTEAN® TGXTM precast protein gels (Bio-Rad, catalog number: 4561084)
Solutions
Buffers for protein purification
1. Luria-Bertani (LB) medium (see Recipes)
2. Yeast-peptone-dextrose (YPD) medium (see Recipes)
3. 1 M TCEP (see Recipes)
4. His column buffer (see Recipes)
5. His wash buffer (see Recipes)
6. His elution buffer (see Recipes)
7. FPLC buffer (see Recipes)
8. TEV cleavage buffer (see Recipes)
Buffers for resin preparation
9. SulfoLink coupling buffer (see Recipes)
10. 50 mM L-cysteine (see Recipes)
Buffers for OtUBD resin-based purifications
11. OtUBD column buffer (see Recipes)
12. OtUBD wash buffer-1 (see Recipes)
13. OtUBD wash buffer-2 (see Recipes)
14. 1× SDS gel sample buffer (see Recipes)
15. OtUBD native lysis buffer (see Recipes)
16. OtUBD urea lysis buffer (see Recipes)
17. OtUBD elution buffer (see Recipes)
18. Neutralization buffer (see Recipes)
19. OtUBD urea wash buffer (see Recipes)
Recipes
Buffers for protein purification
1. Luria-Bertani (LB) medium
Add 5 g of yeast extract, 10 g of tryptone, and 5 g of NaCl to 1 L of MilliQ water.
Autoclave and store at room temperature.
2. Yeast-peptone-dextrose (YPD) medium
Add 10 g of yeast extract and 20 g of peptone to 950 mL of MilliQ water.
Autoclave and then add 2% glucose.
Store at room temperature.
3. 1 M TCEP
Prepare a 1 M stock solution of TCEP and neutralize it to pH 7 with 10 N NaOH.
Store aliquots at -20 °C.
4. His column buffer (for histidine tag purifications)
50 mM Tris-HCl, pH 8.0, 300 mM NaCl, 10 mM imidazole
5. His wash buffer
50 mM Tris-HCl, pH 8.0, 300 mM NaCl, 20 mM imidazole
6. His elution buffer
50 mM Tris-HCl, pH 8.0, 300 mM NaCl, 250 mM imidazole
7. FPLC buffer
50 mM Tris-HCl, 150 mM NaCl pH 7.5, 1 mM TCEP
Prepare fresh. Filter sterilize and purge by applying a vacuum for 20 min.
8. TEV cleavage buffer
50 mM Tris-HCl, pH 8.0, 100 mM NaCl, 1 mM TCEP
Add TCEP right before use.
Buffers for resin preparation
9. SulfoLink coupling buffer
50 mM Tris-HCl, 5 mM EDTA, pH 8.5
10. 50 mM L-cysteine
Dissolve L-cysteine in SulfoLink coupling buffer and adjust the pH to 8.5 using 10 N NaOH.
Prepare fresh before use.
Buffers for OtUBD resin-based purifications
11. OtUBD column buffer
50 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, 0.5% Triton X-100, 10% glycerol, pH 7.5
12. OtUBD wash buffer-1
50 mM Tris-HCl, 150 mM NaCl, 0.05% Tween 20 pH 7.5
13. OtUBD wash buffer-2
50 mM Tris-HCl, 1 M NaCl pH 7.5
14. 1× SDS gel sample buffer
50 mM Tris-HCl pH 6.8, 2% SDS, 5% glycerol, 100 mM DTT, 0.005% bromophenol blue
Aliquot and store at -20 °C.
15. OtUBD native lysis buffer
50 mM Tris-HCl, 300 mM NaCl, 1 mM EDTA, 0.5% Triton X-100, with freshly added 20 mM NEM, cOmplete mini EDTA-free protease inhibitor cocktail (Roche), and 1 mM PMSF, pH 7.5
16. OtUBD urea lysis buffer
50 mM Tris-HCl, 300 mM NaCl, 8 M urea, 1 mM EDTA, 0.5% Triton X-100, with freshly added 20 mM NEM, cOmplete mini EDTA-free protease inhibitor cocktail (Roche), and 1 mM PMSF, pH 7.5. Prepare fresh before use.
17. OtUBD elution buffer
100 mM glycine, pH 2.5 (pH adjusted with HCl)
18. Neutralization buffer
1 M Tris-HCl, pH 9
19. OtUBD urea wash buffer
Dissolve 2.93 g of urea powder in 10 mL of OtUBD native lysis buffer (without adding inhibitors) to make a final concentration of 4 M Urea. Prepare fresh before use.
Note: All the buffers are prepared at room temperature, filter-sterilized, and stored short term (weeks) at room temperature and long term (months) at 4 °C unless otherwise indicated. Filter-sterilization is not needed if all components of the buffer have been kept sterile. (Tris buffers may change in pH depending on the temperature. For the pH 7.5 Tris buffers we prepared at room temperature, the pH falls within the range of 7.5–7.8 at 4 °C.) All urea-containing buffers are prepared fresh before use or stored frozen at -20 °C to minimize urea decomposition. Unless otherwise indicated, all buffers are stable for at least 4 months when stored at 4 °C.
Laboratory supplies
1. Corning® 100 mm TC-treated culture dish (Corning, catalog number: 430167)
2. Corning® 50 mL centrifuge tubes (Millipore Sigma, catalog number: CLS430828-100EA)
3. Corning® 15 mL centrifuge tubes (Millipore Sigma, catalog number: CLS430791-500EA)
4. Amicon® Ultra centrifugal filter, 3 kDa MWCO (Millipore Sigma, catalog number: UFC9003)
5. Corning® 100 mm × 15 mm sterile Petri plates (Millipore Sigma, catalog number: CLS351029)
7. P1000, P200, and P10 pipettes (Pipetman and Eppendorf)
8. Thermo ScientificTM screw cap micro tubes (Thermo Scientific, catalog number: 3488)
9. Porcelain mortar (Capitol Scientific, catalog number: CoorsTek 66310)
10. Porcelain pestle (Capitol Scientific, catalog number: CoorsTek 66311)
11. Pyrex 2,000 mL, No. 4980 Conical flasks
12. 1.5 mL microcentrifuge tubes without caps (USA Scientific Inc., catalog number: NC9380465)
13. Borosil® tubes, test, reusable, plain end, 25 mL (Borosil®)
14. Econo-Pac® chromatography columns (Bio-Rad, catalog number: 7321010)
15. Poly-Prep® chromatography columns (Bio-Rad, catalog number: 7311550)
16. NalgeneTM Rapid-FlowTM sterile disposable bottle top filters (Thermo Scientific, catalog numbers: 595-4520 and 596-3320)
17. Corning® cell lifter (Millipore Sigma, catalog number: CLS3008)
18. InvitrogenTM flat gel loading tips (Fischer Scientific, catalog number: LC1002)
Equipment
1. Ultracold -80 °C freezer (Stirling, USA)
2. Refrigerated incubator shaker (New Brunswick Scientific, model: Innova 4230)
3. Incubator shaker (New Brunswick Scientific, model: Innova 4200)
4. Mini-PROTEANR II Cell (Bio-Rad, USA)
5. Mammalian cell 37 °C incubator with 5% CO2
6. Power pack (VWR, power source 250 V)
7. pH meter instrument (Mettler Toledo, model: SevenDirect SD20)
8. Cold room or 4 °C cabinet setup
9. Rocking shaker (Reliable Scientific, model: 55)
10. Superdex 75 gel filtration column, Hiload 16/600 (Cytiva, catalog number: 28-9893-33)
11. Sorvall Legend X1R centrifuge (Thermo Scientific, catalog number: 335650)
12. Tabletop ultracentrifuge (IEC Micromax, model: OptimaTM L-90K)
13. Mini Vortexer (VWR)
14. Rotoflex R2000 (Argos, China)
15. Misonix Sonicator 3000 with microtip probes
16. Drum roller (New Brunswick, model: TC-7)
17. G: Box imaging system with GeneSnap software
18. E-C Apparatus Corporation EC 105 (37 °C incubator)
19. Low-temperature incubator 815 (30 °C incubator)
20. Lyophilizer (Labconco, USA)
21. Incubator shaker series (New Brunswick Scientific, model: Innova44)
22. BioTek plate reader (Synergy MX)
23. ÄKTA Pure chromatography system GDM-49 (Cytiva, USA)
24. Mettler Toledo Balances (MS 3002S and MS 1045)
25. FastPrep homogenizer (MP Bio, CA, USA)
26. ChemiDoc imaging system (Bio-Rad, USA)
Software and datasets
1. ImageJ v2.14.0/1.54f
2. GraphPad Prism v8.0.1
Procedure
A. Purification of OtUBD
Notes:
1. In this protocol, we describe the purification of recombinant OtUBD with an engineered N-terminal cysteine (Cys-OtUBD) for covalent immobilization on a SulfoLink resin. Covalent immobilization eliminates bait (OtDUB) contamination in eluted proteins and is desired for some downstream applications, such as proteomics.
2. Noncovalent immobilization of maltose-binding protein (MBP)-OtUBD and biotinylated OtUBD have also been successfully applied for ubiquitin substrate enrichment [7,18,19].
3. Two different plasmids are available from our lab (available through Addgene; see Materials and reagents) for expression and purification of Cys-OtUBD. pET21a-cys-His-UBD expresses Cys-OtUBD with an N-terminal His6 tag for easy purification and higher yield. pRT498 expresses a His6-MBP-tagged Cys-OtUBD with a TEV protease-cleavable linker to remove the extra tags. We have used both variants of OtUBD for efficient ubiquitinome and ubiquitin interactome enrichment.
1. Protein expression
a. Transform competent Rosetta (DE3) E. coli with the pET-21a-cys-His-UBD (ampicillin resistance gene) or pRT498-OtUBD (kanamycin resistance gene) plasmid following standard protocols and select single colonies [20].
Note: A small-scale expression test is recommended to ensure the colon(ies) selected express the desired protein before scaling up. Briefly, start a 2 mL overnight culture and dilute the overnight culture by 1:100 in 5 mL of medium as described in step A1b. When the OD600 reaches 0.5–0.8, collect 1 OD600 worth of cells as the uninduced sample by centrifugation at 5,000× g for 5 min and removing the supernatant. Induce the rest of the culture as described in step A1b. The following day, collect 1 OD600 worth of cells as the induced sample. Resuspend the bacteria pellets in 150 μL of 1× SDS sample buffer and boil for 5 min. Load 15 μL of the uninduced and induced samples on an SDS-PAGE gel and stain with GelCode Blue. Cys-His-UBD shows up as an extra band at around 12 kD in the induced samples (Figure 1A, lanes 1 and 2).
b. Start with a 20 mL overnight culture of the transformed E. coli grown at 37 °C in LB supplemented with the appropriate antibiotic (100 μg/mL ampicillin or 50 μg/mL kanamycin). Dilute the overnight culture by 1:100 in 2 L of LB medium supplemented with the corresponding antibiotic and grow the culture in a shaking incubator at 37 °C until the OD600 reaches 0.5–0.8. Protein production is induced by adding IPTG to a final concentration of 0.3 mM. Culture the cells overnight in a shaking incubator at 16–18 °C. The following day, harvest the bacterial cells by centrifugation at 6,000× g for 10 min.
Pause point: The pellet can be processed immediately or stored at -20 °C or below until the next step.
Notes:
1. In our hands, a 2 L E. coli culture yields 6–20 mg of pure Cys-OtUBD.
2. We recommend saving aliquots of uninduced and induced bacteria for SDS-PAGE analysis.
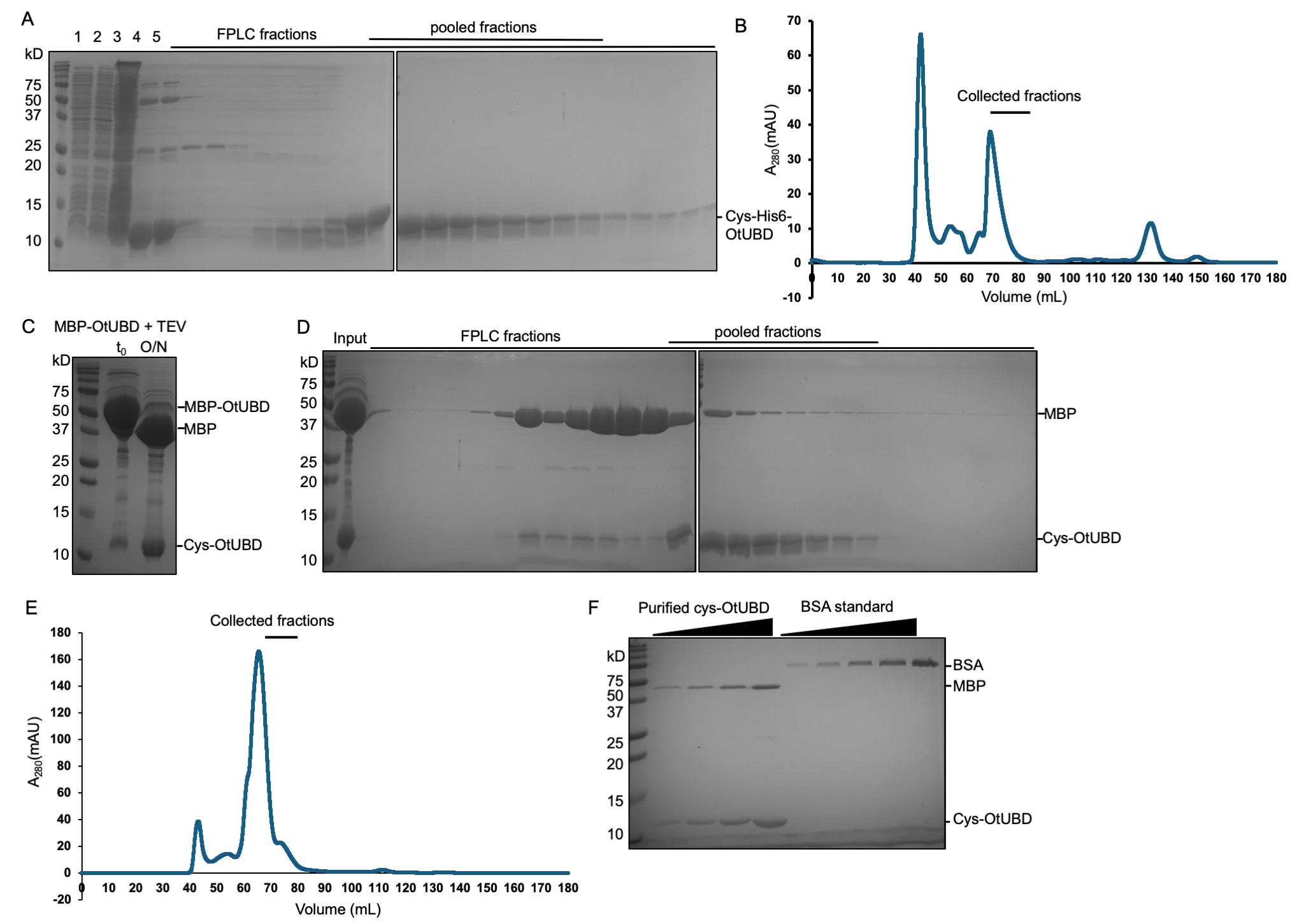
Figure 1. Protein purification of Cys-His6-OtUBD and untagged Cys-OtUBD (from MBP-His6-OtUBD). (A) Image showing the purified Cys-His6-OtUBD protein obtained after size exclusion chromatography (SEC). Lane 1: Uninduced E. coli pellet (15 μL). Lane 2: Induced pellet (15 μL). Lane 3: Lysate (30 μL). Lane 4: Ni-NTA elution (30 μL). Lane 5: Pre-FPLC sample (10 μL). Lane 6–28: FPLC fractions (30 μL). (B) Representative chromatogram of Cys-His6-OtUBD protein in the size exclusion chromatography. (C) Image showing the cleaved Cys-OtUBD obtained before (t0) and after (O/N) TEV protease treatment of 6xHis-MBP-Cys-OtUBD protein (15 μL each). (D) Images of the protein Cys-OtUBD obtained after SEC of TEV-cleaved 6xHis-MBP-Cys-OtUBD (without Ni-NTA cleanup). 10 μL of input and 30 μL of each FPLC fraction were loaded into the gel. (E) Representative chromatogram of Cys-OtUBD protein purification using size exclusion chromatography. (F) Representative SDS-PAGE gel for quantification of untagged Cys-OtUBD. BSA standards: 1:2 serial dilution from 0.125 μg to 2 μg of BSA was loaded from left to right. Purified cys-OtUBD: 1:2 serial dilution from 0.125 μL to 1 μL (volume before diluting in SDS sample buffer) of purified cys-OtUBD was loaded from left to right. All gels in this figure were stained with GelCode Blue.
2. Protein purification of Cys-His-UBD from pET-21a-cys-His-UBD plasmid
a. Resuspend the pellet in 30 mL of bacteria lysis buffer (His column buffer) freshly supplemented with 2 mM PMSF and a pinch of lysozyme and DNase I. Incubate the mixture on ice for 30 min for better lysis efficiency (optional). Bacterial cell lysis can be performed using a sonicator (process time 20–30 min, pulses on for 5 s and off for 5 s) or a French press (1–2 passes).
Note: The bacterial cell lysis should be carried out at 4 °C.
b. Clarify the lysate by centrifuging at 10,000× g for 1 h or at 45,000× g for 25 min at 4 °C, saving the supernatant (pH of the supernatant should be 7–8.5 for optimal binding to the Ni-NTA column).
c. Keeping all reagents and equipment at 4 °C, if possible, transfer 2 mL of Ni-NTA resin (4 mL of 1:1 slurry) to a disposable gravity column, drain the storage buffer, and equilibrate the resin by passing through 25 mL of His column buffer. Incubate the clarified lysate with the equilibrated resin in the sealed column for 1 h with end-over-end rotation.
d. Secure the column in an upright position and allow the resin bed to settle. Drain the supernatant and wash the resin with 25 mL of His column buffer, followed by 25 mL of His wash buffer. Elute the Cys-His-UBD by passing 8 mL of His elution buffer through the resin and collecting the eluate in a clean 15 mL tube. Add 5 mM TCEP reducing agent to the eluate immediately.
e. Fractionate the Ni-NTA column eluate by FPLC using an ÄKTA Pure chromatography system with HiLoad 16/600 Superdex 75 column (Cytiva) or similar (Figure 1A, B). Fractionation should be done at 4 °C using FPLC buffer (see Recipes). Pool and concentrate the fractions containing pure OtUBD as determined by SDS-PAGE and protein staining. Aliquot the purified protein, flash-freeze the aliquots with liquid nitrogen, and store at -80 °C.
Pause point: The protein can be stored for a long time without any degradation.
Notes:
1. We recommend running aliquots of the input, flowthrough, both wash fractions, and the eluted fraction from the Ni-NTA pulldown on an SDS-PAGE for quality control and troubleshooting, especially for the first time purifying this protein.
2. TCEP helps prevent intermolecular disulfide formation between the added cysteinyl group in OtUBD molecules and is compatible at this concentration with the subsequent SulfoLink coupling. If DTT is used instead in the eluted fractions, it must be removed before the SulfoLink coupling step.
3. The 8 mL eluate can be concentrated down to 2–3 mL in an Amicon filtration unit for loading on the size-exclusion column step (A2e) Alternatively, the eluate can be collected in 1 mL fractions in Eppendorf tubes, and the two fractions with the highest protein concentration identified by BCA assay or similar can be directly loaded; the remaining eluate fractions can be combined and concentrated to 2 mL and then run on the size-exclusion column.
4. We recommend proceeding to the FPLC purification step immediately. Leaving the eluate at 4 °C for a prolonged time may lead to the formation of precipitates, which need to be removed before loading on the size-exclusion column.
5. OtUBD has no tyrosine or tryptophan residues; it has a zero predicted extinction coefficient at A280 nm and may not show up in the A280 nm UV trace. We recommend running FPLC fractions on an SDS-PAGE gel and determining the concentration of purified OtUBD by SDS-PAGE and protein staining or a colorimetric assay such as the BCA assay using bovine serum albumin (BSA) standard.
3. Protein purification of untagged Cys-OtUBD
a. Express and purify 6xHis-MBP-Cys-OtUBD expressed in E. coli from the pRT498 plasmid following the instructions in steps A1 and step A2a–d. Following elution from Ni-NTA resin, buffer exchange the eluate with TEV buffer (see Recipes) using either a desalting column or a 3000 Da cutoff centrifugal filter following the manufacturer’s instructions. If using a centrifugal filter, concentrate the solution and dilute with TEV buffer. Repeat these two steps several times until the final concentration of imidazole is less than 20 mM. (Note: The concentration can be estimated with the volumes before and after each dilution.) Estimate the concentration of crude 6xHis-MBP-Cys-OtUBD with A280 nm reading or BCA assay and adjust the final concentration to less than 20 mg/mL.
b. Add approximately 1:80 (w/w) His-tagged TEV protease to 6xHis-MBP-Cys-OtUBD and incubate overnight without shaking at 4 °C. (Note: The mass of the protein is calculated from the estimated concentration of MBP-UBD.) Check cleavage efficiency by running aliquots taken before and after incubation with TEV on an SDS-PAGE gel (Figure 1C). Optional: Post incubation, pass the mixture through a column with Ni-NTA resin to capture the cleaved tag and TEV protease. Wash the resin with 10 mL of His wash buffer and combine the two flowthrough fractions.
Notes:
1. We typically use a final concentration of around 100 μg/mL for TEV protease. We have not tested lower concentrations of TEV protease. We recommend a small-scale testing if lower concentrations of TEV protease are to be used.
2. The Ni-NTA resin used for the purification of the 6xHis-MBP-Cys-OtUBD can be reused here after washing with 5 column volumes of 0.5 M NaOH and 10 column volumes of His column buffer.
c. Size fractionate the Ni-NTA column eluate by FPLC using an ÄKTA Pure chromatography system with HiLoad 16/600 Superdex 75 column (Cytiva) or similar (Figure 1D, E). Fractionation should be done at 4 °C using the FPLC buffer (see Recipes). Pool and concentrate the fractions containing OtUBD as determined by SDS-PAGE and protein staining (Figure 1F). Aliquot the purified protein, flash-freeze the aliquots with liquid nitrogen, and store at -80 °C.
Notes:
1. Cleaved MBP may not be completely removed during the Ni-NTA capture and FPLC steps. However, as MBP contains no cysteine residues, this contamination will not affect the SulfoLink coupling step. We recommend determining the concentration of purified OtUBD by SDS-PAGE gel against a serial dilution of BSA standards. BCA or Bradford assays are not suitable due to the MBP contamination (Figure 1F).
2. The yield of untagged Cys-OtUBD purified by this procedure is around 1 mg per 2 L of bacterial culture, significantly lower than Cys-His-OtUBD.
B. OtUBD-resin preparation
1. Coupling OtUBD to the SulfoLink resin
a. Bring SulfoLink coupling resin (Thermo Scientific) to room temperature. Transfer 2 mL (bed volume) of resin to a 20 mL gravity column. Let the resin settle and drain the storage buffer. Pass 8 mL of SulfoLink coupling buffer (4 bed volumes) through the resin to equilibrate it.
Note: SulfoLink coupling resin is provided as a 50% slurry, and care should be taken to suspend the resin evenly by swirling the bottle before taking out the resin. A wide-mouthed pipette should be used to transfer the resin.
b. Dilute 4 mg of the purified Cys-OtUBD (His tagged or untagged) protein (obtained from section A) in 4 mL of coupling buffer freshly supplemented with 20 mM TCEP and incubate for 30 min at room temperature. Cap the bottom of the column, add the Cys-OtUBD solution to the resin, secure the top cap, and mix with end-over-end rotation for 30–60 min at room temperature. (Note: Cys-OtUBD efficiently couples with the resin in 30 min, whereas Cys-His6-OtUBD requires a 60 min incubation time for full coupling.) Let the resin settle. Collect the flowthrough and wash the resin with 6 mL of coupling buffer.
c. To block any remaining free active sites on the resin, react the resin with 4 mL of freshly prepared 50 mM L-cysteine (see Recipes) [21], incubating for 30 min at room temperature with gentle rotation. Let the resin settle before draining the liquid. Wash the resin by passing through 12 mL of 1 M NaCl, followed by 4 mL of OtUBD column buffer. For storage, add 2 mL of OtUBD column buffer containing 0.05% NaN3 to form a 50% slurry and keep the resin at 4 °C.
Notes:
1. The coupling efficiency of the resin can be estimated by checking the amount of OtUBD in the input and flowthrough fraction collected after incubation (Figure 2A, B).
2. Keep the resin-containing tube in an upright position to prevent it from drying. We have used the resin for at least 1.5 years after preparation without significant loss of efficacy.
Caution: At no point during the procedure should the resin bed be allowed to dry.
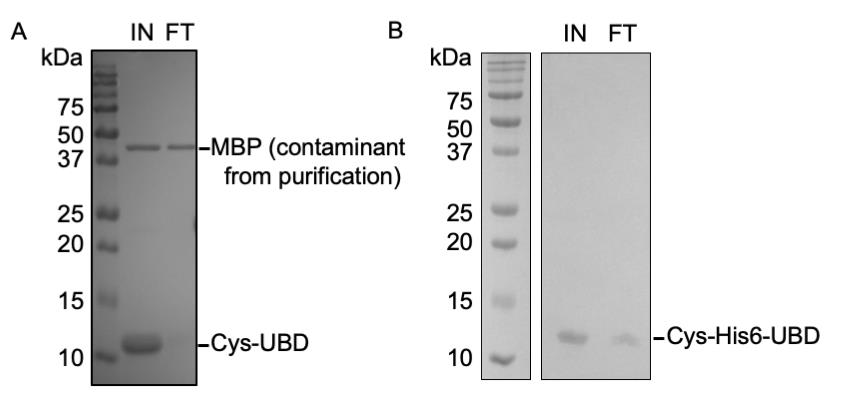
Figure 2. Resin binding efficiencies of the Cys-OtUBD and Cys-His6-OtUBD proteins. (A, B) GelCode Blue staining of the gel comparing the amount of Cys-OtUBD (A) and Cys-His6-OtUBD (B) protein in the sample before and after incubation with the Sulfolink coupling resin. IN: Input; FT: flowthrough.
2. Preparation of Cys-coupled resin (negative control resin)
To make the Sulfolink resin capped with cysteine, follow steps B1a and then B1c.
C. Biological sample collection
1. Saccharomyces cerevisiae cell collection
a. We recommend starting with a minimum of 50 OD600 equivalents of yeast cells (e.g., 50 mL of yeast culture at an OD600 of 1) to provide sufficient lysate input. If downstream analysis is proteomics, we recommend starting with 1 L of yeast culture (if harvested at around 1 OD600) per sample.
b. After the desired cell growth period and possible culture treatments, harvest the cells by centrifugation at 5,000× g for 10 min at 4 °C. After discarding the supernatant, resuspend the cell pellet in pre-chilled MilliQ water and centrifuge again as above. The supernatant is discarded, and the pellet is flash frozen in liquid nitrogen. The pellet can be stored at -80 °C until further use.
Note: We have used nutrient-rich YPD media as well as selective minimal media for growing the yeast cells, and affinity purifications are highly reproducible with either.
2. Mammalian cell collection
a. We recommend starting with a minimum of one 10-cm dish of confluent adherent mammalian cells to provide sufficient input for the pulldown. If downstream analysis involves proteomics, we recommend starting with five 10-cm dishes per sample.
b. After the desired period of cell culture and potential treatment, remove the medium from the adherent cell monolayers. Wash the cells once with cold DPBS before dislodging the cells by scraping with a cell lifter in 1 mL of cold DPBS on ice.
c. Transfer the dislodged cells to an appropriate tube and pellet them by centrifugation at 400× g for 4 min. Remove the supernatant by careful aspiration. The cell pellets can be flash frozen in liquid nitrogen and stored at -80 °C until used.
Note: We have tested this protocol with HeLa cells with or without the treatment of 4-NQO with success. We recommend that the reader performs a small-scale experiment first to determine the right scale for their own system.
D. OtUBD resin-based purification of ubiquitin conjugates and binding proteins under native conditions: preparation of lysates and affinity purification procedures
1. Preparation of yeast lysate
Method 1: Cryogrinding
a. Pre-chill a mortar and pestle with liquid nitrogen. Transfer the frozen cell pellet to the pre-cooled mortar. (The tube containing the frozen pellet can be carefully broken by a mallet. Transfer the pieces of the pellet into the mortar with a clean pair of forceps.) Carefully press the pellet with the pestle to break it down into smaller pieces. Then grind the frozen yeast (with liquid nitrogen in the mortar bowl) into a fine powder with fast circular motions.
Notes:
1. This method is suitable for larger-scale cell extractions (> 50 mL of yeast culture at 1 OD600). Choose the size of the mortar and pestle according to the number of yeast cells.
2. To pre-chill the mortar and pestle, add liquid nitrogen to the mortar and completely submerge the tip of the pestle in liquid nitrogen. Initially, the liquid nitrogen will boil and evaporate fast. Repeatedly add liquid nitrogen until the “boiling” slows down. At this point, the frozen cell pellet can be transferred to the mortar. Protect the benchtop by setting the mortar on top of a layer of Styrofoam (e.g., lid of a Styrofoam box).
Caution: Keep adding liquid nitrogen to the mortar throughout the grinding process. Do not let the liquid nitrogen run out for more than 30 s.
Notes:
1. Care should be taken to avoid spilling of powdered cells during grinding.
2. The finished yeast powder resembles a fine flour. Grinding may take more than 10 min for each sample.
b. Carefully transfer the powdered yeast to a 15 mL plastic conical tube pre-chilled in liquid nitrogen.
Caution: Do not allow the yeast powder to thaw at any stage, as it allows protein degradation.
Pause point: The yeast powder can be stored at -80 °C before starting with the next step.
c. To 1 volume of yeast powder, add 1 volume of ice-cold OtUBD native lysis buffer. Incubate the mixture on ice with intermittent vortexing every 3–4 min for 10 min.
d. Clarify the crude lysate by centrifugation at 21,000× g for 10–12 min at 4 °C. Carefully transfer the supernatant to a pre-cooled fresh tube.
e. Measure lysate protein concentration with BCA or Bradford assay and adjust the concentration to 2–4 mg/mL if possible.
Method 2: Bead beating
a. For smaller-scale purifications (50–100 mL of yeast cultures at 1 OD600), a simple bead-beating method can be employed to extract proteins. Resuspend cell pellets in 1 mL of cold OtUBD native lysis buffer supplemented with 10% glycerol and 0.6 mL glass beads and transfer to a chilled screw cap microcentrifuge tube. Perform the lysis process at 4 °C using a FastPrep homogenizer with the following cycle conditions: set speed at 5.0 m/s, 3 pulses of 30 s bead beating with 1 min rest on ice in between, 4 min rest on ice, another 3 pulses of 30 s bead beating with 1 min rest on ice in between.
b. Incubate the mixture for 5 min on ice and then centrifuge at 8,000× g for 5 min at 4 °C. Transfer the supernatant (crude lysate) to a pre-chilled Eppendorf tube. Rinse the beads and pelleted debris with 0.5 mL of cold lysis buffer, centrifuge at 8,000× g for 5 min at 4 °C, and add the supernatant to the first collected fraction. Centrifuge the combined supernatants at 21,000× g for 12 min at 4 °C. Transfer the cleared lysate to a pre-chilled Eppendorf tube. Measure lysate protein concentration with BCA or Bradford assay and adjust the concentration to 2–4 mg/mL if possible.
2. Preparation of mammalian cell lysates
a. Resuspend cell pellets (obtained as described in section C2) in ice-cold native lysis buffer (add 0.25–0.5 mL of lysis buffer per 10-cm cell dish). Incubate the mixture on ice with intermittent vortexing every 5–8 min for 30–40 min.
b. Centrifuge the lysate at 21,000× g for 20 min and transfer the clarified lysate to a pre-chilled fresh Eppendorf tube.
c. Measure lysate protein concentration with BCA or Bradford assay and adjust the concentration to 2–4 mg/mL if possible.
3. Affinity purifications using the OtUBD resin
We recommend starting with 25 μL of resin (bed volume) per 1 mg of lysate protein. We have had success using as little as 25 μL of total resin for western blotting and 0.25–2 mL of resin for proteomics experiments.
a. Affinity purifications suitable for medium-to-large scale “native” purifications (requires >250 μL of resin bed volume): The procedure will enrich both ubiquitinated proteins and ubiquitin-binding proteins.
i. Transfer the appropriate amount of OtUBD resin to a 10 or 20-mL gravity column and allow the resin to settle for 10 min.
ii. Drain the excess resin storage buffer and equilibrate the resin with 5 bed volumes of OtUBD column buffer. Alternatively, wash the resin with 2 bed volumes of OtUBD elution buffer, followed by 20 bed volumes of OtUBD column buffer. This washing procedure helps further eliminate contamination by OtUBD that had not been covalently linked to the resin beads, which can be helpful for applications such as proteomics.
Note: Make sure enough OtUBD column buffer is passed through the resin immediately after the wash with OtUBD elution buffer to neutralize the acidic pH.
iii. Add the cleared mammalian or yeast cell lysate to the resin and cap the column on both ends. Incubate the mixture for 2 h with gentle end-over-end rotation at 4 °C to allow binding between the lysate and the resin.
iv. Allow the resin to settle in the column for 10 min and then collect the unbound lysate as flowthrough. Wash the resin by passing through OtUBD column buffer (15 bed volumes), followed by OtUBD wash buffer-1 (15 bed volumes), and then OtUBD wash buffer-2 (15 bed volumes). Perform this step at 4 °C if possible.
v. The elution process varies depending on the downstream process, as follows:
Elution by SDS denaturation: Incubate the resin with 2–3 resin bed volumes of 1× SDS gel sample buffer (see Recipes) at room temperature for 15 min with end-over-end rotation. If using a column for elution, drain the eluate into an appropriately sized tube(s). If using a microcentrifuge tube for the elution step, centrifuge and collect the supernatant.
Notes:
1. Resin cannot be reused after this elution condition.
2. This elution method is more efficient than low pH elution but may result in the elution of some proteins that nonspecifically stick to the resin.
Elution by low pH: Wash the resin with 2 bed volumes of MilliQ water to push out the remaining buffer and then elute the bound proteins by resuspending the resin in 2 bed volumes of OtUBD elution buffer and incubating with gentle rotation for 5 min at 4 °C. Collect the eluate and immediately neutralize it by adding 0.2 bed volumes of neutralization buffer. Repeat this step one to two times to ensure complete protein elution.
Notes:
1. We recommend saving an aliquot of the eluate(s) for pulldown quality control and protein concentration estimation (see next section).
2. This elution method allows re-using the affinity resin. To regenerate the resin, following acid elution, wash by passing 5 additional bed volumes of OtUBD elution buffer, followed by 20 bed volumes of OtUBD column buffer through the resin. Make sure the buffer coming out of the resin is no longer acidic toward the end of the column buffer wash. For storage, pass 2 bed volumes of OtUBD column buffer containing 0.05% NaN3 (storage buffer), cap the bottom of the column, and add 1 bed volume of this storage buffer to form a 50% slurry. Keep the column tightly sealed at 4 °C. We have successfully reused the resin without significant loss of efficiency up to two times.
3. To reduce total volume and salt content for LC–MS/MS sample preparation, flash freeze the eluate in liquid nitrogen and freeze-dry in a lyophilizer. Reconstitute the lyophilized eluates in water and perform a standard methanol-chloroform extraction [22].
b. Affinity purifications suitable for smaller-scale “native” purifications (<200 μL resin bed volume): The procedure will enrich both ubiquitinated proteins and ubiquitin-binding proteins.
i. Transfer an appropriate amount of OtUBD resin to a microcentrifuge tube and centrifuge at 1,000× g for 2 min. Carefully remove the supernatant by pipetting or vacuum aspiration. Equilibrate the resin, incubate with lysate, wash the resin, and elute as described under step D3a, except that after each step, collect or remove the supernatant carefully by pipetting after beads are pelleted by centrifugation (1,000× g for 2 min).
Caution! Extra care should be taken to remove as much supernatant as possible without removing any resin, as resin loss will greatly increase variability across samples. Use fine pipette tips, such as flat gel loading tips, with vacuum or manual pipetting to avoid accidental removal of resin. Aim the tip against the wall of the tube and avoid touching the resin bed directly.
E. OtUBD resin-based purification of ubiquitin conjugates and binding proteins from denatured extracts: preparation of lysates and affinity purification procedures
1. (Option 1) Denatured lysates can be prepared as in step D1 or D2, except substituting the OtUBD native lysis buffer with OtUBD urea lysis buffer (see Recipes). Resuspend yeast powder or mammalian cell frozen pellet in OtUBD urea lysis buffer and incubate at room temperature for 15 min with vortexing about every 5 min. Clarify the crude lysate by centrifugation. For bead beating, incubate at room temperature for 5 min after homogenization before centrifugation.
Note: Compared to Option 2, this method could, in theory, include insoluble ubiquitinated proteins and extract more ubiquitinated proteins (e.g., see Figure 4E, D1 vs. D2 from Zhang et al. [18]).
2. (Option 2) Alternatively, denatured lysate can be prepared by adding urea powder to a clarified native lysate (prepared as in step D1 or D2) to a final concentration of 8 M (3.245 g urea/4 mL of lysate). Allow urea to completely dissolve by vortexing and inverting the tube at room temperature. When the urea is completely dissolved in the lysate, incubate the lysate at 25 °C for 30 min.
Note: Adding solid urea will increase the total volume of the final lysate. A native lysate at 12.584 mg/mL protein concentration will result in an 8 M urea lysate at approximately 8 mg/mL [23].
3. Following incubation of the lysate in 8 M urea, bring the urea concentration down to 4 M by addition of a volume of native lysate buffer equal to the volume of the 8 M urea-containing lysate. Keep the mixture on ice from this step on. (Optional) Estimate the total protein content using the BCA or Bradford assay. If possible, use 2–4 mg/mL protein concentration.
Note: When the concentration of urea is diluted to 4 M by the addition of native lysis buffer, most proteins remain denatured, but ubiquitin can fold back to its native state. This allows the binding of ubiquitin moieties to the OtUBD resin in subsequent steps and helps eliminate proteins that are not covalently ubiquitinated (see Figure 3 from [18]).
4. Perform the affinity purification as described in step D3, except that the first wash step is done with OtUBD urea wash buffer instead of OtUBD column buffer (see Recipes).
Note: Urea can decompose into cyanate in aqueous solution, which can covalently modify proteins and lead to artifacts in subsequent proteomic analyses. Make sure all urea-containing buffers are prepared fresh, especially if the downstream applications include proteomic analysis.
F. Analysis of affinity purification quality with immunoblotting and SYPRO Ruby staining
1. Anti-ubiquitin immunoblotting
We recommend performing an anti-ubiquitin immunoblot to determine the efficiency of ubiquitin and ubiquitin-conjugate purification and for gross comparison of ubiquitin-protein profiles in extracts from different cells or conditions.
a. Prepare gel loading samples of the input fraction, unbound protein fraction, (optional) wash fraction(s), and eluate(s) by directly mixing the appropriate amount of 4× SDS sample buffer and heating the samples at 100 °C for 5 min.
b. Resolve the protein samples on a gradient SDS-PAGE gel such as a 4%–15% precast Tris-glycine polyacrylamide gel or similar. We typically do a wet transfer of proteins from the gel to an Immobilon-P (Millipore) PVDF membrane at 70 V for 2.5 h at 4 °C using a Bio-Rad transfer apparatus.
Notes:
1. When performing immunoblotting [18], it is helpful to load normalized ratios of input, flowthrough, and eluate fractions. For example, if the total volume of the eluate is 1/10th of the input lysate volume, and 10 μL of the elution sample is taken to prepare the sample for gel loading, then 1 μL of the input should be used for comparison. Sample volumes should be equalized with 1× SDS sample buffer.
2. We have successfully performed western blots with multiple anti-ubiquitin antibodies and nonfat milk as the blocking agent.
2. SYPRO Ruby gel analysis for total protein visualization and quantification
Total protein visualization in the eluents can also be useful for quality control and troubleshooting. Due to the low abundance of ubiquitinated species in biological samples, we recommend using the highly sensitive SYPRO Ruby protein stain (Invitrogen) to visualize and quantify total protein in the eluates. Perform the SYPRO Ruby staining process according to the manufacturer’s protocol and image the gel on a ChemiDoc imaging system (Bio-Rad) or similar. For protein quantification, use serial dilutions of input lysate of known total protein concentration as the standard (start from 1:100 dilution) (see Figure S3C and S5B from [18]) and quantify lane intensities of the image with ImageJ software.
Interpretation of results: In a ubiquitin immunoblot of a typical OtUBD-based purification, the elution fraction should show enrichment of ubiquitinated proteins, while the unbound protein fraction shows the depletion of ubiquitinated proteins compared to the normalized input lane (e.g., Figure 2B from [18]). Pulldowns under native conditions enrich both ubiquitylated proteins (and free ubiquitin) as well as ubiquitin-interacting proteins. However, pulldowns from denatured lysates should enrich only covalently modified proteins. Indeed, comparison of purifications from native and denatured lysates shows fewer proteins under the latter conditions (see Figure 3H from [18]).
Validation of protocol
This protocol and variations of it have been used and validated in the following research articles:
• Zhang et al. [18]. A versatile new tool derived from a bacterial deubiquitylase to detect and purify ubiquitylated substrates and their interacting proteins. PLoS Biology (original publication)
• Wendrich et al. [7]. Discovery and mechanism of K63-linkage-directed deubiquitinase activity in USP53. Nature Chemical Biology (Figure 2)
• Kazi et al. [19]. Chimeric deubiquitinase engineering reveals structural basis for specific inhibition of the mitophagy regulator USP30. Nature Structural & Molecular Biology (Figure 5)
• Körner et al. [24]. p97/VCP is required for piecemeal autophagy of aggresomes. Nature Communications (Figure 6)
• Banerjee et al. [25]. Light-Activatable Ubiquitin for Studying Linkage-Specific Ubiquitin Chain Formation Kinetics. Advanced Science (Supplementary Figure 3)
• Ojeda-Naharros et al. [26]. Tonic ubiquitination of the central body weight regulator melanocortin receptor 4 (MC4R) promotes its constitutive exit from cilia. PLOS Biology (Figure 7)
General notes and troubleshooting
General notes
1. This protocol aims to provide a general ubiquitin pulldown method suitable for various situations and downstream applications. Variations of this protocol have also been described by other groups [7,19,24–26]. We recommend that readers choose between different options we and others have described, according to the specific system under study and the biological question to address.
2. We recommend performing a small-scale pulldown experiment when first trying out the protocol or a test run with a mock sample. Follow the steps laid out in section F to assess the efficiency of the pulldown.
3. We recommend a small-scale test run if changes are to be made to the conditions that have been tested.
Troubleshooting
Problem 1: Precipitation appears in the 8M urea-containing lysate.
Possible causes: The sample was kept at a low temperature (e.g., on ice) for too long, and urea crashes out; too much urea was added.
Solutions: Warm the lysate up to room temperature and mix thoroughly by inverting the tube several times; double-check the calculation to make sure the final concentration of urea does not exceed 8 M.
Problem 2: Low binding efficiency to the OtUBD resin (high ubiquitin level in the flowthrough).
Possible causes: Sample concentration is too low; not enough resin was used; ubiquitin species does not refold properly after dilution of the 8 M urea-containing lysate.
Solutions: Increase sample concentration; increase incubation time with the resin; use more resin; dilute urea-containing lysate in native lysis buffer so that the final concentration of urea is 4 M or less.
Problem 3: Low elution efficiency with OtUBD elution buffer.
Possible cause: Proteins precipitate during the pulldown steps; we have noticed that the elution efficiency is slightly lower for pulldown under denatured conditions than native conditions.
Solution: Make sure the resin does not dry out at any point during the pulldown; try eluting at room temperature or 37 °C; use SDS sample buffer for elution (additional SDS removal steps are needed if SDS is not compatible with the downstream application).
Problem 4: Lower than expected levels of ubiquitination are detected on the protein of interest.
Possible cause: The ubiquitinated species may be insoluble in the native lysis buffer.
Solution: Try extracting proteins from the samples directly in urea lysis buffer (Section E, Option 1).
Problem 5: A band close to the expected molecular weight of the unmodified protein of interest shows up in the western blot of the eluate of OtUBD pulldown under native conditions.
Possible cause: The unmodified protein of interest might associate with ubiquitin or a ubiquitinated protein; for a larger protein of interest, it may be hard to distinguish the unmodified protein from its monoubiquitinated species due to the small molecular weight difference.
Solution: Compare pulldown results under native and denatured conditions. If the band only shows up under native conditions, it is likely the unmodified protein of interest co-purifying with ubiquitin or a ubiquitinated protein; if the band shows up similarly in both conditions, it is likely to be a monoubiquitinated species of the protein of interest.
Acknowledgments
Conceptualization, M.Z., N.S., M.H.; Investigation, M.Z., N.S.; Writing, N.S., M.Z.; Writing—Review & Editing, M.Z., M.H., N.S.; Funding acquisition, M.H.; Supervision, M.H. Funding supporting this work was provided by NIH grant R35 GM136325 to M.H. The Graphical overview was created with Biorender.com (https://BioRender.com/8lwj041). This protocol is a more detailed and modified version of the earlier published protocol by Zhang et al. [18].
Competing interests
The authors declare that they have no competing interests.
References
- Pickart, C. M. (2001). Mechanisms underlying ubiquitination. Annu Rev Biochem. 70: 503–533. https://doi.org/10.1146/annurev.biochem.70.1.503
- van Wijk, S. J. and Timmers, H. T. (2010). The family of ubiquitin-conjugating enzymes (E2s): deciding between life and death of proteins. FASEB J. 24(4): 981–993. https://doi.org/10.1096/fj.09-136259
- Zheng, N. and Shabek, N. (2017). Ubiquitin Ligases: Structure, Function, and Regulation. Annu Rev Biochem. 86: 129–157. https://doi.org/10.1146/annurev-biochem-060815-014922
- Otten, E. G., Werner, E., Crespillo-Casado, A., Boyle, K. B., Dharamdasani, V., Pathe, C., Santhanam, B. and Randow, F. (2021). Ubiquitylation of lipopolysaccharide by RNF213 during bacterial infection. Nature. 594(7861): 111–116. https://doi.org/10.1038/s41586-021-03566-4
- Komander, D. and Rape, M. (2012). The ubiquitin code. Annu Rev Biochem. 81: 203–229. https://doi.org/10.1146/annurev-biochem-060310-170328
- Grice, G. L. and Nathan, J. A. (2016). The recognition of ubiquitinated proteins by the proteasome. Cell Mol Life Sci. 73(18): 3497–3506. https://doi.org/10.1007/s00018-016-2255-5
- Wendrich, K., Gallant, K., Recknagel, S., Petroulia, S., Kazi, N. H., Hane, J. A., Fuhrer, S., Bezstarosti, K., O'Dea, R., Demmers, J., et al. (2025). Discovery and mechanism of K63-linkage-directed deubiquitinase activity in USP53. Nat Chem Biol. 21(5): 746–757. https://doi.org/10.1038/s41589-024-01777-0
- Senft, D., Qi, J. and Ronai, Z. A. (2018). Ubiquitin ligases in oncogenic transformation and cancer therapy. Nat Rev Cancer. 18(2): 69–88. https://doi.org/10.1038/nrc.2017.105
- Zheng, Q., Huang, T., Zhang, L., Zhou, Y., Luo, H., Xu, H. and Wang, X. (2016). Dysregulation of Ubiquitin-Proteasome System in Neurodegenerative Diseases. Front Aging Neurosci. 8: 303. https://doi.org/10.3389/fnagi.2016.00303
- Newton, K., Matsumoto, M. L., Ferrando, R. E., Wickliffe, K. E., Rape, M., Kelley, R. F. and Dixit, V. M. (2012). Using linkage-specific monoclonal antibodies to analyze cellular ubiquitylation. Methods Mol Biol. 832: 185–196. https://doi.org/10.1007/978-1-61779-474-2_13
- Matsumoto, M. L., Dong, K. C., Yu, C., Phu, L., Gao, X., Hannoush, R. N., Hymowitz, S. G., Kirkpatrick, D. S., Dixit, V. M. and Kelley, R. F. (2012). Engineering and structural characterization of a linear polyubiquitin-specific antibody. J Mol Biol. 418(3–4): 134–144. https://doi.org/10.1016/j.jmb.2011.12.053
- Fulzele, A. and Bennett, E. J. (2018). Ubiquitin diGLY Proteomics as an Approach to Identify and Quantify the Ubiquitin-Modified Proteome. Methods Mol Biol. 1844: 363–384. https://doi.org/10.1007/978-1-4939-8706-1_23
- Ellison, M. and Hochstrasser, M. (1991). Epitope-tagged ubiquitin. A new probe for analyzing ubiquitin function. J Biol Chem. 266(31): 21150–21157. https://doi.org/10.1016/s0021-9258(18)54833-x
- Hjerpe, R., Aillet, F., Lopitz-Otsoa, F., Lang, V., England, P. and Rodriguez, M. S. (2009). Efficient protection and isolation of ubiquitylated proteins using tandem ubiquitin-binding entities. EMBO Rep. 10(11): 1250–1258. https://doi.org/10.1038/embor.2009.192
- Kaiser, S. E., Riley, B. E., Shaler, T. A., Trevino, R. S., Becker, C. H., Schulman, H. and Kopito, R. R. (2011). Protein standard absolute quantification (PSAQ) method for the measurement of cellular ubiquitin pools. Nat Methods. 8(8): 691–696. https://doi.org/10.1038/nmeth.1649
- McDowell, G. S. and Philpott, A. (2013). Non-canonical ubiquitylation: mechanisms and consequences. Int J Biochem Cell Biol. 45(8): 1833–1842. https://doi.org/10.1016/j.biocel.2013.05.026
- Berk, J. M., Lim, C., Ronau, J. A., Chaudhuri, A., Chen, H., Beckmann, J. F., Loria, J. P., Xiong, Y. and Hochstrasser, M. (2020). A deubiquitylase with an unusually high-affinity ubiquitin-binding domain from the scrub typhus pathogen Orientia tsutsugamushi. Nat Commun. 11(1): 2343. https://doi.org/10.1038/s41467-020-15985-4
- Zhang, M., Berk, J. M., Mehrtash, A. B., Kanyo, J. and Hochstrasser, M. (2022). A versatile new tool derived from a bacterial deubiquitylase to detect and purify ubiquitylated substrates and their interacting proteins. PLoS Biol. 20(6): e3001501. https://doi.org/10.1371/journal.pbio.3001501
- Kazi, N. H., Klink, N., Gallant, K., Kipka, G. M. and Gersch, M. (2025). Chimeric deubiquitinase engineering reveals structural basis for specific inhibition of the mitophagy regulator USP30. Nat Struct Mol Biol. https://doi.org/10.1038/s41594-025-01534-4
- Sambrook, J. and Russell, D. W. (2003). Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory. ISBN: 9780879695774.
- Domen, P. L., Nevens, J. R., Mallia, A. K., Hermanson, G. T. and Klenk, D. C. (1990). Site-directed immobilization of proteins. J Chromatogr. 510: 293–302. https://doi.org/10.1016/s0021-9673(01)93763-x
- Wessel, D. and Flugge, U. I. (1984). A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal Biochem. 138(1): 141–143. https://doi.org/10.1016/0003-2697(84)90782-6
- Wingfield, P. T. (2001). Use of protein folding reagents. Curr Protoc Protein Sci. Appendix 3: Appendix 3A. https://doi.org/10.1002/0471140864.psa03as00
- Körner, M., Muller, P., Das, H., Kraus, F., Pfeuffer, T., Spielhaupter, S., Oeljeklaus, S., Schulein-Volk, C., Harper, J. W., Warscheid, B., et al. (2025). p97/VCP is required for piecemeal autophagy of aggresomes. Nat Commun. 16(1): 4243. https://doi.org/10.1038/s41467-025-59556-x
- Banerjee, S., Cakil, Z. V., Gallant, K., Boom, J. V. D., Palei, S., Meyer, H., Gersch, M. and Summerer, D. (2025). Light-Activatable Ubiquitin for Studying Linkage-Specific Ubiquitin Chain Formation Kinetics. Adv Sci (Weinh). 12(6): e2406570. https://doi.org/10.1002/advs.202406570
- Ojeda-Naharros, I., Das, T., Castro, R. A., Bazan, J. F., Vaisse, C. and Nachury, M. V. (2025). Tonic ubiquitination of the central body weight regulator melanocortin receptor 4 (MC4R) promotes its constitutive exit from cilia. PLoS Biol. 23(2): e3003025. https://doi.org/10.1371/journal.pbio.3003025
Article Information
Publication history
Received: Jun 6, 2025
Accepted: Jul 22, 2025
Available online: Aug 6, 2025
Published: Sep 5, 2025
Copyright
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Saha, N., Zhang, M. and Hochstrasser, M. (2025). Use of a High-Affinity Ubiquitin-Binding Domain to Detect and Purify Ubiquitinated Substrates and Their Interacting Proteins. Bio-protocol 15(17): e5426. DOI: 10.21769/BioProtoc.5426.
Category
Biochemistry > Protein > Immunodetection > Western blot
Systems Biology > Proteomics
Biochemistry > Protein > Modification
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Tips for asking effective questions
+ Description
Write a detailed description. Include all information that will help others answer your question including experimental processes, conditions, and relevant images.
Share
Bluesky
X
Copy link








