- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Production of Homogeneous, Functional Zinc-Finger Arrays in High Yield With Two Chromatographic Steps
(§Technical contact: liangjch@stanford.edu) Published: Vol 15, Iss 16, Aug 20, 2025 DOI: 10.21769/BioProtoc.5420 Views: 1477
Reviewed by: Navnita DuttaAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Optimized Secretome Sample Preparation From High Volume Cell Culture Media for LC–MS/MS Proteomic Analysis
Basil Baby Mattamana [...] Peter Allen Faull
Dec 20, 2025 1261 Views

A One-Step Method for Efficient Purification of Functional Cas9 Protein
Xinzhi Duan [...] Aihua Mao
Feb 5, 2026 158 Views

On-Column Dual-Gradient Refolding for Efficient Recovery of Insoluble Affinity-Tagged Recombinant Proteins
Anna Vlaskina [...] Maxim Patrushev
Feb 5, 2026 67 Views
Abstract
Zinc-finger (ZF) arrays are compact, sequence-specific polynucleotide-binding domains, which have been used to target the delivery of diverse effector domains, enabling applications such as gene identification, localization, regulation, and editing. To facilitate in vitro applications of ZF arrays, we have developed a general method for their expression and purification. Here, we describe a protocol involving two chromatographic steps that yields homogeneous and functional ZF arrays in milligram quantities.
Key features
• A general method for expressing and purifying C2H2 ZF arrays in E. coli, compatible with both natural and artificial ZFs.
• The His-SUMO tag improves ZF-array solubility, eliminating the need for denaturation and refolding steps.
• Simple, two-step purification yields milligram-scale ZF arrays suitable for downstream applications.
Keywords: Zinc fingersGraphical overview
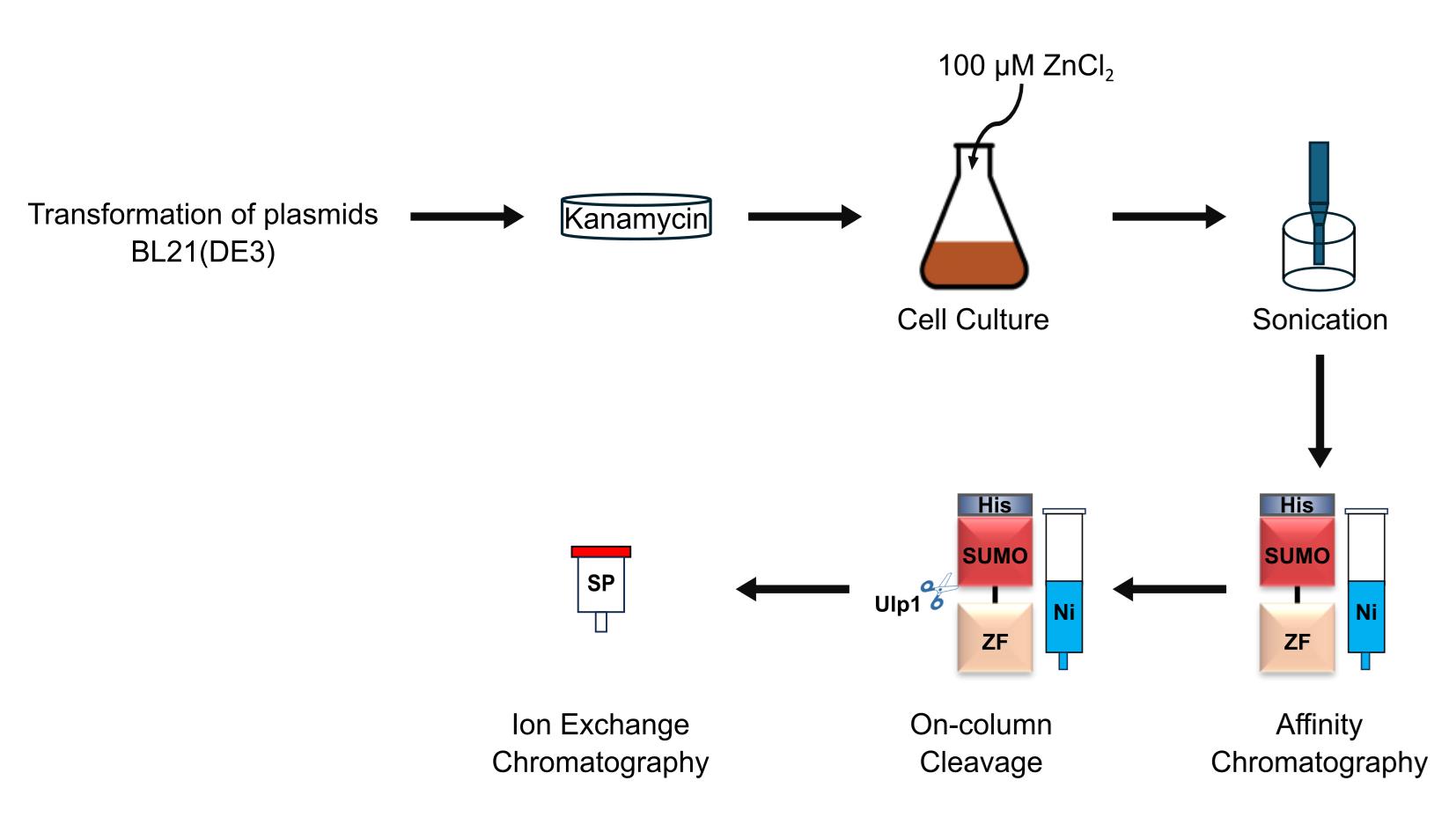
Background
Zinc-finger (ZF) proteins—initially identified in Xenopus laevis transcription factor IIIA [1]—are DNA-binding domains found in the most prevalent family of transcription factors in metazoans [2–4]. ZFs adopt a variety of structural motifs, among which the most abundant and well-studied is that of the C2H2 family, which comprises between 28 and 30 amino acids, including two cysteine and two histidine residues that tetrahedrally coordinate the central zinc ion [5,6]. The “recognition helix” of the resulting ββα fold projects into the major groove of DNA, forming hydrogen bonds with three consecutive base pairs [7,8]. The recognition of extended sequences is made possible by the assembly of ZFs in tandem to form polydactyl arrays [6,9–14]. Because the specificity of each finger is affected by its neighbors [9], the development of polydactyl arrays has posed considerable challenges. Recently, these challenges have been mitigated by the advent of deep-learning methods, enabling the design of ZF arrays targeting arbitrary DNA sequences [15].
ZF arrays are commonly expressed directly in target cells by the introduction of transgenic expression constructs [16–20]. An alternative approach, involving purified ZF arrays, has demonstrated certain advantages, such as greater efficiencies and lower off-target rates in gene editing by ZF nucleases [21]. However, the production of ZF arrays in suitable amounts and purity for exogenous delivery has been hitherto challenging, as there has been a need to optimize the expression and purification procedures for each new ZF array [22–30]. Existing strategies include the use of an affinity tag—polyhistidine (His-tag) [22–24], glutathione S-transferase (GST) [25,26], or maltose-binding protein (MBP) [27,28]—and the exploitation of biochemical properties of ZFs, such as their coordination to zinc ions or their specific binding to DNA sequences [29,30]. To avoid the requirement of tailoring procedures to each unique ZF array, we developed a simple and general method, based on the His-SUMO tag, for the expression and purification of functional ZF arrays in milligram quantities [31]. We present the procedure here in full detail.
Materials and reagents
A. Expression of ZF arrays in E. coli BL21 (DE3)
1. pET-28a-SUMO-ZF plasmids (Genewiz)
2. QIAprep® Spin Miniprep kit (QIAGEN, catalog number: 27104)
3. BL21 (DE3) competent E. coli (New England Biolabs, catalog number: C2527H)
4. Tryptone (Fisher Scientific, catalog number: BP14212)
5. Yeast extract (USBiological, catalog number: Y2008)
6. Kanamycin sulfate (Teknova, catalog number: K2150)
7. Agar (Fisher Scientific, catalog number: A360-500)
8. Sodium chloride (NaCl) (Fisher Scientific, catalog number: BP358-212)
9. Zinc chloride (ZnCl2) (Sigma-Aldrich, catalog number: Z0152)
10. Isopropyl β-D-1-thiogalactopyranoside (IPTG) (GOLDBIO, catalog number: I2481C100)
B. Purification of ZF arrays in E. coli BL21 (DE3)
1. Phenylmethylsulfonyl fluoride (PMSF) (Sigma-Aldrich, catalog number: P7626-250MG)
2. Benzamidine hydrochloride monohydrate (GOLDBIO, catalog number: B-050-500)
3. Pepstatin A (GOLDBIO, catalog number: P-020-100)
4. Leupeptin hemisulfate (GOLDBIO, catalog number: L-010-100)
5. Absolute ethanol (Fisher Scientific, catalog number: BP2818-4)
6. Ni-NTA Superflow (QIAGEN, catalog number: 1018611)
7. PierceTM Bradford Plus Protein Assay Reagent (Thermo Scientific, catalog number: 23238)
8. HiTrapTM SP HP column (Cytiva, catalog number: 17115101)
9. Tris base (Fisher Scientific, catalog number: BP1521)
10. Hydrochloric acid (HCl) ~37% (Thermo Scientific, catalog number: LC149501)
11. Imidazole (Sigma-Aldrich, catalog number: 56750-500g)
12. 2-Mercaptoethanol (MP, catalog number: 190242)
13. L-Arginine (Sigma-Aldrich, catalog number: 11009-100G)
14. Coomassie Brilliant Blue R-250 Dye (Thermo Scientific, catalog number: 20278)
15. 2-Propanol (J.T. Baker, catalog number: 9084-01)
16. Acetic acid (Fisher Scientific, catalog number: A38SI-212)
17. NuPAGETM Bis-Tris Mini Protein Gels, 4%–12%, 1.0–1.5 mm (Invitrogen, catalog number: NP0321BOX)
18. 2-(N-Morpholino)ethanesulfonic acid (MES) (Sigma-Aldrich, catalog number: M3671)
19. Sodium dodecyl sulfate (SDS) (Invitrogen, catalog number: 15525-017)
20. Ethylenediaminetetraacetic acid (EDTA) (Fisher Scientific, catalog number: BP120-1)
21. Precision Plus ProteinTM Dual Color Standards (Bio-Rad, catalog number: 1610374)
22. Ulp1 (home-made, Addgene plasmid #229224) or commercial SUMO Protease (Invitrogen, catalog number: 12588-018)
Solutions
1. LB medium (see Recipes)
2. LB agar (see Recipes)
3. Kanamycin stock (1,000×) (see Recipes)
4. ZnCl2 stock (0.1 M) (see Recipes)
5. IPTG stock (1 M) (see Recipes)
6. PMSF stock (100×) (see Recipes)
7. Protease inhibitor cocktail stock (200×) (see Recipes)
8. Imidazole stock, pH 7.5 (5 M) (see Recipes)
9. L-Arginine stock (1M) (see Recipes)
10. NaCl stock (5 M) (see Recipes)
11. Tris-HCl, pH 7.5 (1 M) (see Recipes)
12. Coomassie Brilliant Blue R-250 staining solution (see Recipes)
13. Lysis buffer (see Recipes)
14. Wash buffer (see Recipes)
15. Cleavage buffer A (see Recipes)
16. Cleavage buffer B (see Recipes)
17. Stock buffer (see Recipes)
18. 20× MES SDS running buffer (see Recipes)
Recipes
1. LB medium
| Reagent | Final concentration | Amount |
|---|---|---|
| Tryptone | 10 g/L | 10 g |
| Yeast extract | 5 g/L | 5 g |
| Sodium chloride | 10 g/L | 10 g |
| Double-distilled water (ddH2O) | N/A | Up to 1 L |
Autoclave the LB medium and store at room temperature.
2. LB agar plates
| Reagent | Final concentration | Amount |
|---|---|---|
| Tryptone | 10 g/L | 10 g |
| Yeast extract | 5 g/L | 5 g |
| Sodium chloride | 10 g/L | 10 g |
| Agar | 15 g/L | 15 g |
| ddH2O | N/A | Up to 1 L |
Autoclave the prepared LB Agar. Allow it to cool to approximately 50 °C, then add antibiotics, mix thoroughly, and pour plates immediately. Store the prepared LB agar plates inverted at 4 °C.
3. Kanamycin stock (1,000×)
| Reagent | Final concentration | Amount |
|---|---|---|
| Kanamycin sulfate | 50 mg/mL | 2.5 g |
| ddH2O | N/A | Up to 50 mL |
Sterilize the solution by passing it through a 0.22 μm polyethersulfone filter and store at -20 °C.
4. ZnCl2 stock (0.1 M)
| Reagent | Final concentration | Amount |
|---|---|---|
| Zinc chloride | 0.1 M | 13.628 g |
| ddH2O | N/A | Up to 1 L |
If the powder does not dissolve completely after stirring, add a few drops of concentrated HCl (~37%) while stirring to facilitate dissolution. Sterilize the solution by passing it through a 0.22 μm polyethersulfone filter and store at room temperature.
5. IPTG stock (1 M)
| Reagent | Final concentration | Amount |
|---|---|---|
| IPTG | 1 M | 11.915 g |
| ddH2O | N/A | Up to 50 mL |
Sterilize the solution by passing through a 0.22 μm polyethersulfone filter and store at -20 °C.
6. PMSF stock (100×)
| Reagent | Final concentration | Amount |
|---|---|---|
| PMSF | 100 mM | 1.7419 g |
| Absolute ethanol | N/A | Up to 100 mL |
Store at -20 °C.
7. Protease inhibitor cocktail stock (200×)
| Reagent | Final concentration | Amount |
|---|---|---|
| Benzamidine hydrochloride monohydrate | 0.4 M | 3.1322 g |
| Pepstatin A | 0.4 mM | 13.7178 mg |
| Leupeptin hemisulfate | 0.12 mM | 2.8536 mg |
| Absolute ethanol | N/A | Up to 50 mL |
Store at -20 °C.
8. Imidazole stock, pH 7.5 (5 M)
| Reagent | Final concentration | Amount |
|---|---|---|
| Imidazole | 5 M | 340.4 g |
| HCl | N/A | Adjust pH to 7.5 |
| ddH2O | N/A | Up to 1 L |
Wrap the bottle with aluminum foil to protect from light and store at room temperature.
9. L-Arginine stock (1 M)
| Reagent | Final concentration | Amount |
|---|---|---|
| L-Arginine | 1 M | 174.2 g |
| HCl | N/A | Adjust pH to 7.5 |
| ddH2O | N/A | Up to 1 L |
Store at room temperature.
10. NaCl stock (5 M)
| Reagent | Final concentration | Amount |
|---|---|---|
| Sodium chloride | 5 M | 292.2 g |
| ddH2O | N/A | Up to 1 L |
Store at room temperature.
11. Tris-HCl, pH 7.5 (1 M)
| Reagent | Final concentration | Amount |
|---|---|---|
| Tris base | 1 M | 121.14 g |
| HCl | N/A | Adjust pH to 7.5 |
| ddH2O | N/A | Up to 1 L |
Store at room temperature.
12. Coomassie Brilliant Blue R-250 staining solution
| Reagent | Final concentration | Amount |
|---|---|---|
| Coomassie Brilliant Blue R-250 Dye | 1 g/L | 1 g |
| 2-Propanol | N/A | 250 mL |
| Acetic acid | N/A | 100 mL |
| ddH2O | N/A | Up to 1 L |
Mix completely and store at room temperature.
13. Lysis buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| Tris-HCl, pH 7.5 (1 M) | 20 mM | 20 mL |
| NaCl stock (5 M) | 2 M | 400 mL |
| Imidazole stock, pH 7.5 (5 M) | 10 mM | 2 mL |
| ZnCl2 stock (0.1 M) | 100 μM | 1 mL |
| ddH2O | N/A | Up to 1 L |
Store at 4 °C.
14. Wash buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| Tris-HCl, pH 7.5 (1 M) | 20 mM | 20 mL |
| NaCl stock (5 M) | 500 mM | 100 mL |
| Imidazole stock, pH 7.5 (5 M) | 60 mM | 12 mL |
| ZnCl2 stock (0.1 M) | 100 μM | 1 mL |
| ddH2O | N/A | Up to 1 L |
Store at 4 °C.
15. Cleavage buffer A
| Reagent | Final concentration | Amount |
|---|---|---|
| Tris-HCl, pH 7.5 (1 M) | 20 mM | 20 mL |
| NaCl stock (5 M) | 100 mM | 20 mL |
| ZnCl2 stock (0.1 M) | 100 μM | 1 mL |
| L-Arginine stock (1M) | 100 mM | 100 mL |
| 2-Mercaptoethanol | 5 mM | 340 μL |
| ddH2O | N/A | Up to 1 L |
Prepare the solution freshly before use and store at 4 °C.
16. Cleavage buffer B
| Reagent | Final concentration | Amount |
|---|---|---|
| Tris-HCl, pH 7.5 (1 M) | 20 mM | 20 mL |
| NaCl stock (5 M) | 1 M | 200 mL |
| ZnCl2 stock (0.1 M) | 100 μM | 1 mL |
| L-Arginine stock (1 M) | 100 mM | 100 mL |
| 2-Mercaptoethanol | 5 mM | 340 μL |
| ddH2O | N/A | Up to 1 L |
Prepare the solution freshly before use and store at 4 °C.
17. Stock buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| Tris-HCl, pH 7.5 (1 M) | 20 mM | 20 mL |
| NaCl stock (5 M) | 100 mM | 20 mL |
| ZnCl2 stock (0.1 M) | 100 μM | 1 mL |
| L-Arginine stock (1 M) | 100 mM | 100 mL |
| ddH2O | N/A | Up to 1 L |
Store at 4 °C.
18. 20× MES SDS running buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| MES | 1 M | 97.6 g |
| Tris base | 1 M | 60.6 g |
| SDS | 69.3 mM | 10 g |
| EDTA | 20.5 mM | 3.8 g |
| ddH2O | N/A | Up to 500 mL |
Store at room temperature. The pH of 1× MES SDS running buffer should be 7.3.
Laboratory supplies
1. Thermo Scientific NalgeneTM Rapid-FlowTM sterile disposable bottle-top filters with PES, CN, SFCA or Nylon membranes (Fisher Scientific, catalog number: 09-741-07)
2. Petri dish, untreated, UltraCruz® (Santa Cruz Biotechnology, catalog number: sc-516726)
3. FalconTM 50 mL high-clarity conical centrifuge tubes (Fisher Scientific, catalog number: 14-432-22)
4. Pyrex® narrow-mouth graduated Erlenmeyer flask (Millipore, catalog number: CLS498050-12EA)
5. Borosilicate glass narrow-mouth Erlenmeyer flasks, 3,000 mL (United Scientific, catalog number: FG4980-3000)
6. Econo-Pac® chromatography columns (Bio-Rad, catalog number: 7321010)
7. Amicon® Ultra centrifugal filter, 3 kDa MWCO (Millipore, catalog number: UFC9003)
Equipment
1. FisherbrandTM IsotempTM general purpose deluxe water baths (Fisher Scientific, catalog number: FSGPD02)
2. Digital incubator (VWR, model: 1525)
3. Refrigerated centrifuge (Eppendorf, model: 5810R)
4. Refrigerated incubator shaker (New Brunswick Scientific, model: Innova 4430)
5. Sonicator (QSONICA, model: Q500)
6. SorvallTM LYNX 6000 superspeed centrifuge (Thermo Scientific, catalog number: 75006591)
7. Roto-Torque® heavy duty rotator (Cole-Parmer, model: 7637)
8. Econo pump (Bio-Rad, model: EP-1)
9. ÄKTA pureTM micro (Cytiva, catalog number: 29302479)
10. XCell SureLockTM Mini-Cell (Invitrogen, catalog number: EI0001)
11. ChemiDocTM MP imaging system (Bio-Rad, catalog number: 12003154)
12. NanoDropTM 2000c Spectrophotometer (Thermo Scientific, catalog number: ND-2000C)
13. FiberliteTM F9-6 × 1000 LEX fixed angle rotor for LYNX 6000 superspeed centrifuge (Thermo Scientific, catalog number: 096-061075)
14. FiberliteTM F21-8 × 50y fixed angle rotor with auto-lock for LYNX 4000 and 6000 superspeed centrifuges (Thermo Scientific, catalog number: 096-084275)
Software and datasets
1. UnicornTM 7 for ÄKTA purifier (GE Healthcare, https://www.cytivalifesciences.com/en/us/shop/chromatography/software/unicorn-7)
Procedure
A. Transformation of E. coli BL21 (DE3) with ZF-array expression plasmids
1. DNA fragments encoding various ZF arrays were optimized for E. coli codon usage, synthesized, and cloned by Genewiz in a pET-28a-SUMO vector using BamHI and XhoI restriction sites. The constructed plasmids (pET-28a-SUMO-ZF, where “ZF” is a placeholder for a particular ZF array) were delivered as E. coli agar stab cultures, and plasmids were prepared therefrom by alkaline lysis and silica column chromatography (e.g., Qiagen Miniprep Kit).
2. Thaw 50 μL of competent E. coli BL21 (DE3) cells on ice.
3. Add 2 μL of the pET-28a-SUMO-ZF plasmid (~30 ng/μL) to the competent cells. Mix by gently flicking the tube.
4. Incubate the competent cells on ice for 30 min, then heat-shock in a water bath (e.g., FisherbrandTM IsotempTM water bath) at 42 °C for 45 s. Place the cells on ice for an additional 3 min.
5. Add 500 μL of LB medium to the competent cells and shake at 180 rpm for 50 min at 37 °C (e.g., Innova 4430 shaker).
6. Spread 200 μL of the transformed cells on 10 cm LB agar plates containing kanamycin (final concentration: 50 μg/mL).
7. Incubate the plates (inverted) overnight at 37 °C (e.g., VWR 1525 incubator).
8. The next day, pick a single colony from the plate using a sterile pipette tip and inoculate into a 50 mL flask containing 15 mL of LB medium supplemented with kanamycin (final concentration: 50 μg/mL).
9. Grow the starter culture overnight at 37 °C at 180 rpm (e.g., Innova 4430 shaker).
B. Expression of ZF arrays (1 L culture)
1. Prepare 1 L of LB medium in a 3 L shaker flask and autoclave. After cooling, add kanamycin to 50 μg/mL (1 mL of 1,000× stock) and ZnCl2 to 100 μM (1 mL of 0.1 M stock).
2. Add 15 mL of starter culture to the 1 L of LB medium and grow at 37 °C in a shaker at 180 rpm (e.g., Innova 4430 shaker).
3. Monitor the OD600 of the culture; when it reaches ~0.7, reduce the temperature to 16 °C and induce the expression of ZF arrays by adding IPTG to 0.3 mM (300 μL of 1 M stock).
4. Incubate the culture overnight (~16 h) at 16 °C while shaking at 180 rpm.
5. Harvest the cells by centrifuging at 4,000× g for 20 min at 4 °C (e.g., LYNX 6000 centrifuge with an F9–6×1000 rotor).
6. Discard the supernatant and flash-freeze the cell pellet in liquid nitrogen.
Note: The frozen pellet can be stored at -80 °C for at least one year.
C. Purification of ZF arrays
Day 1. ZF-array purification—affinity chromatography: Ni-NTA
1. Pour 2 mL of Ni-NTA Superflow resin slurry (50% v/v resin) into a gravity-flow chromatography column (e.g., Econo-Pac® Chromatography Column).
2. Wash the resin with 10 column volumes each of ddH2O and lysis buffer.
3. Thaw the frozen cell pellet on ice and resuspend it in lysis buffer (10 mL of lysis buffer per gram of cell pellet) supplemented with 1× PMSF (100 μL of 100× stock solution per 10 mL of lysis buffer) and 1× Protease inhibitor cocktail (50 μL of 200× stock solution per 10 mL of lysis buffer).
4. Lyse the resuspended cells on ice with a tip sonicator (e.g., Q500 Sonicator. Amplitude: 50%; 15 s on/45 s off; 5 min).
Note: We recommend the use of type-304 stainless-steel beakers during sonication due to their high thermal conductivity and cavitation erosion resistance.
5. Centrifuge the lysate at 60,000× g for 1 h at 4 °C (e.g., LYNX 6000 centrifuge with an F21–8×50y rotor).
6. Apply the clear supernatant to the packed bed of Ni-NTA Superflow resin.
7. Wash the resin with lysis buffer while monitoring the protein concentration of the flowthrough by mixing 10 μL of the column effluent with 50 μL of Bradford reagent and observing the color (or measuring the absorbance at 595 nm). Continue washing until there is no detectable protein in the effluent.
8. Wash the resin with wash buffer until there is no detectable protein in the effluent.
9. Wash the resin with cleavage buffer A until there is no detectable protein in the effluent.
Note: All washing steps (steps C7–9) are typically performed using 10–20 column volumes of the corresponding buffer.
10. Resuspend the resin in 5 mL of cleavage buffer A.
Note: We suggest using five bed volumes of cleavage buffer A; larger volumes may reduce cleavage efficiency.
11. Add 0.1 mg of homemade Ulp1 protease to the resuspended resin for on-column cleavage of the His-SUMO tag.
Notes:
1. N-terminally His-tagged Ulp1 was expressed in E. coli BL21 (DE3) and purified by Ni-NTA affinity chromatography, following the same procedure used for ZF-array expression and purification. After loading the clarified supernatant onto the Ni-NTA Superflow resin, the resin was washed with wash buffer. Ulp1 was subsequently eluted using wash buffer containing 300 mM imidazole. The eluted Ulp1 was concentrated to 1 mg/mL and mixed with glycerol to a final concentration of 50% (v/v). The final Ulp1 was flash-frozen in liquid nitrogen and stored at -80 °C.
2. Homemade Ulp1 can be replaced with a commercial Ulp1 (SUMO Protease, catalog number: 12588-018).
12. Seal the column and incubate on a rotator at 4 °C overnight (e.g., Cole-Parmer rotator).
Day 2. ZF-array purification—cation exchange chromatography: Sulfopropyl-Sepharose
1. Equilibrate the HiTrap SP HP column with five column volumes of ddH2O using a peristaltic pump at a flow rate of 1 mL/min (e.g., EP-1 pump).
2. Equilibrate the column with five column volumes of cleavage buffer A using the peristaltic pump at a flow rate of 1 mL/min.
3. Collect the cleaved ZF arrays from the Ni-NTA Superflow resin (flowthrough fraction) in a 50 mL Falcon tube.
4. Wash the Ni-NTA Superflow resin with additional cleavage buffer A and collect the flowthrough fraction until there is no detectable protein in the effluent.
5. Mix the two flowthrough fractions and centrifuge them at 4,000× g for 20 min at 4 °C to remove insoluble material.
6. Load the clarified supernatant onto the HiTrap SP HP column at a flow rate of 1 mL/min using the peristaltic pump.
7. Flush the ÄKTA pureTM micro system with cleavage buffer A and then insert the HiTrap SP HP column into its flow path.
Note: Before starting the run, configure the ÄKTA system to continually monitor UV absorbance at 280 nm (A280), 254 nm (A254), and 215 nm (A215).
8. Wash the column at a flow rate of 1 mL/min with cleavage buffer A until all absorbance curves reach a stable baseline.
9. Elute the ZF arrays using a linear gradient from 0% to 100% cleavage buffer B over 60 min. Collect the eluate in 1-mL fractions.
Note: For most ZF arrays, elution can be monitored by A280. However, for ZF arrays with low molar absorptivity at 280 nm (e.g., due to a lack of tryptophan residues), the presence of ZF arrays can be monitored by the absorbance at 215 nm. Typically, only one elution peak is observed in either the A280 or A215 signal.
10. Analyze the eluate fractions by SDS-PAGE. Stain the gel with Coomassie Brilliant Blue R-250 staining solution, and then destain with water until a clear background is obtained.
11. Concentrate the fractions containing pure ZF arrays using a centrifugal filter device with an appropriate molecular-weight cutoff (e.g., Amicon® Ultra Centrifugal Filter, 3 kDa MWCO) while exchanging the buffer to stock buffer.
12. (Optional) Perform preparative gel-filtration chromatography to ensure monodispersity of size.
Note: If this step is performed, the buffer exchange described in step 11 above can be omitted.
13. Determine the concentration of the purified ZF array by measuring the absorption at 280 nm (e.g., using a NanoDrop spectrophotometer). Aliquot and flash-freeze the protein solution in liquid nitrogen and then store at -80 °C.
Notes:
1. For ZF arrays with very low molar absorptivity at 280 nm, the protein concentration can instead be measured according to the method of Bradford [32].
2. Frozen ZF arrays remain stable at -80 °C for at least 6 months. For optimal protein stability, we recommend avoiding repeated freeze-thaw cycles.
Validation of protocol
This protocol has been used and validated in the following research article:
• Liang et al. [31]. A universal method for the purification of C2H2 zinc finger arrays. PLOS One (Figures 1E, 2, and 3A, top panel).
General notes and troubleshooting
General notes
1. This protocol has been tested on ZF arrays with isoelectric points (pI) ranging from 7.8 to 10.6 [31]. For ZF arrays with a pI at or below 7.5, the pH of the buffers may need to be adjusted accordingly to ensure a net positive charge in solution.
2. Beginning with Section C, all steps should be carried out on ice or in a cold room.
3. Due to the presence of 2-mercaptoethanol in cleavage buffer A, we recommend using fresh Ni-NTA Superflow resin.
4. Zinc ions should be present throughout the entire expression and purification process. L-Arginine should be included in all steps starting from Ulp1 cleavage.
Troubleshooting
Problem 1: After overnight Ulp1 treatment, cleaved ZF arrays fail to elute from Ni-NTA Superflow resin.
Possible cause: ZF arrays may exhibit strong, nonspecific binding to the resin.
Solution: Elute ZF arrays with imidazole-containing wash buffer.
Problem 2: Multiple bands, at or above the expected MW, appear on SDS-PAGE gels of purified ZF arrays.
Possible cause: Upon denaturation, cysteine residues in ZF domains may form intermolecular disulfide bonds, leading to bands corresponding to monomers, dimers, or higher-order complexes.
Solution: Add fresh 2-mercaptoethanol to the protein loading buffer and use freshly prepared samples.
Problem 3: Bands below the expected MW appear on SDS-PAGE gels of purified ZF arrays.
Possible cause: Protease contamination.
Solution: Add PMSF/protease inhibitor cocktail to purification buffers immediately before use.
Problem 4: Cleaved His-SUMO tag and Ulp1 appear in the eluate.
Possible cause: The binding affinity of Ni-NTA Superflow resin may be reduced by the inclusion of 2-mercaptoethanol.
Solution: Use fresh resin. The His-SUMO tag and Ulp1 can also be removed by the HiTrap SP HP column step.
Problem 5: Incomplete cleavage of the His-SUMO tag.
Possible cause: The activity of the batch of homemade Ulp1 may be suboptimal, or the cleavage duration may be insufficient.
Solution: Use a new batch of Ulp1 or increase its concentration. Extending the duration of cleavage may also be helpful.
Problem 6: Low protein expression after induction.
Possible cause: Poor protein expression.
Solution: If, after repeating the transformation, expression continues to be low, optimize the IPTG concentration and the induction temperature. If necessary, redesign the construct (ensuring bacterial codon optimization).
Problem 7: Protein expression observed before IPTG induction.
Possible cause: Basal expression from the T7 promoter.
Solution: Use the BL21 (DE3) pLysS (or pLysE) strain instead of BL21 (DE3) to suppress basal expression.
Acknowledgments
R.D.K., A.J.B., and P.J.M. conceived and initiated the project. P.J.M., A.J.B., M.A., R.D.K., and Y.N. supervised the work. J.L. and G.W. performed the experiments. J.L. drafted the initial manuscript. P.J.M., A.J.B., and Y.N. revised the manuscript.
We thank Dr. Jia Liu for providing the initial ZF arrays plasmids. We thank Ms. Xiuxia Gao and Ms. Feifan Wen for their contributions during the initial stage of this research.
This research was supported by funds from private sources (for R.D.K), NIH grants T32GM007365 and 1DP5OD033431 (for A.J.B.), and a grant from ShanghaiTech University (for Y.N.).
This protocol was described and validated in Liang et al. [31].
Competing interests
The authors declare no conflicts of interest.
References
- Miller, J., McLachlan, A. and Klug, A. (1985). Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 4(6): 1609–1614. https://doi.org/10.1002/j.1460-2075.1985.tb03825.x
- Emerson, R. O. and Thomas, J. H. (2009). Adaptive Evolution in Zinc Finger Transcription Factors. PLos Genet. 5(1): e1000325. https://doi.org/10.1371/journal.pgen.1000325
- Tupler, R., Perini, G. and Green, M. R. (2001). Expressing the human genome. Nature. 409(6822): 832–833. https://doi.org/10.1038/35057011
- Najafabadi, H. S., Garton, M., Weirauch, M. T., Mnaimneh, S., Yang, A., Kim, P. M. and Hughes, T. R. (2017). Non-base-contacting residues enable kaleidoscopic evolution of metazoan C2H2 zinc finger DNA binding. Genome Biol. 18(1): 167. https://doi.org/10.1186/s13059-017-1287-y
- Klug, A. and Schwabe, J. (1995). Zinc fingers: Protein Motifs 5. FASEB J. 9(8): 597–604.
- Klug, A. (2010). The Discovery of Zinc Fingers and Their Applications in Gene Regulation and Genome Manipulation. Annu Rev Biochem. 79(1): 213–231. https://doi.org/10.1146/annurev-biochem-010909-095056
- Pavletich, N. P. and Pabo, C. O. (1991). Zinc Finger-DNA Recognition: Crystal Structure of a Zif268-DNA Complex at 2.1 Å. Science. 252(5007): 809–817. https://doi.org/10.1126/science.2028256
- Fairall, L., Schwabe, J. W. R., Chapman, L., Finch, J. T. and Rhodes, D. (1993). The crystal structure of a two zinc-finger peptide reveals an extension to the rules for zinc-finger/DNA recognition. Nature. 366(6454): 483–487. https://doi.org/10.1038/366483a0
- Choo, Y. and Klug, A. (1994). Toward a code for the interactions of zinc fingers with DNA: selection of randomized fingers displayed on phage. Proc Natl Acad Sci USA. 91(23): 11163–11167. https://doi.org/10.1073/pnas.91.23.11163
- Wu, H., Yang, W. P. and Barbas, C. F. (1995). Building zinc fingers by selection: toward a therapeutic application. Proc Natl Acad Sci USA. 92(2): 344–348. https://doi.org/10.1073/pnas.92.2.344
- Bae, K. H., Do Kwon, Y., Shin, H. C., Hwang, M. S., Ryu, E. H., Park, K. S., Yang, H. Y., Lee, D. k., Lee, Y., Park, J., et al. (2003). Human zinc fingers as building blocks in the construction of artificial transcription factors. Nat Biotechnol. 21(3): 275–280. https://doi.org/10.1038/nbt796
- Isalan, M., Klug, A. and Choo, Y. (2001). A rapid, generally applicable method to engineer zinc fingers illustrated by targeting the HIV-1 promoter. Nat Biotechnol. 19(7): 656–660. https://doi.org/10.1038/90264
- Magnenat, L., Blancafort, P. and Barbas, C. F. (2004). In Vivo Selection of Combinatorial Libraries and Designed Affinity Maturation of Polydactyl Zinc Finger Transcription Factors for ICAM-1 Provides New Insights into Gene Regulation. J Mol Biol. 341(3): 635–649. https://doi.org/10.1016/j.jmb.2004.06.030
- Gupta, A., Christensen, R. G., Rayla, A. L., Lakshmanan, A., Stormo, G. D. and Wolfe, S. A. (2012). An optimized two-finger archive for ZFN-mediated gene targeting. Nat Methods. 9(6): 588–590. https://doi.org/10.1038/nmeth.1994
- Ichikawa, D. M., Abdin, O., Alerasool, N., Kogenaru, M., Mueller, A. L., Wen, H., Giganti, D. O., Goldberg, G. W., Adams, S., Spencer, J. M., et al. (2023). A universal deep-learning model for zinc finger design enables transcription factor reprogramming. Nat Biotechnol. 41(8): 1117–1129. https://doi.org/10.1038/s41587-022-01624-4
- Mendenhall, E. M., Williamson, K. E., Reyon, D., Zou, J. Y., Ram, O., Joung, J. K. and Bernstein, B. E. (2013). Locus-specific editing of histone modifications at endogenous enhancers. Nat Biotechnol. 31(12): 1133–1136. https://doi.org/10.1038/nbt.2701
- Beerli, R. R., Segal, D. J., Dreier, B. and Barbas, C. F. (1998). Toward controlling gene expression at will: Specific regulation of the erbB-2/HER-2 promoter by using polydactyl zinc finger proteins constructed from modular building blocks. Proc Natl Acad Sci USA. 95(25): 14628–14633. https://doi.org/10.1073/pnas.95.25.14628
- Kim, Y. G., Cha, J. and Chandrasegaran, S. (1996). Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci USA. 93(3): 1156–1160. https://doi.org/10.1073/pnas.93.3.1156
- Gordley, R. M., Smith, J. D., Gräslund, T. and Barbas, C. F. (2007). Evolution of Programmable Zinc Finger-recombinases with Activity in Human Cells. J Mol Biol. 367(3): 802–813. https://doi.org/10.1016/j.jmb.2007.01.017
- Lindhout, B. I., Fransz, P., Tessadori, F., Meckel, T., Hooykaas, P. J. and van der Zaal, B. J. (2007). Live cell imaging of repetitive DNA sequences via GFP-tagged polydactyl zinc finger proteins. Nucleic Acids Res. 35(16): e107–e107. https://doi.org/10.1093/nar/gkm618
- Liu, J., Gaj, T., Wallen, M. C. and Barbas, C. F. (2015). Improved Cell-Penetrating Zinc-Finger Nuclease Proteins for Precision Genome Engineering. Mol Ther Nucleic Acids. 4: e232. https://doi.org/10.1038/mtna.2015.6
- Song, Y., Cui, C., Zhu, H., Li, Q., Zhao, F. and Jin, Y. (2015). Expression, purification and characterization of zinc-finger nuclease to knockout the goat beta-lactoglobulin gene. Protein Expression Purif. 112: 1–7. https://doi.org/10.1016/j.pep.2015.04.004
- Chant, A., Kraemer-Pecore, C. M., Watkin, R. and Kneale, G. G. (2005). Attachment of a histidine tag to the minimal zinc finger protein of the Aspergillus nidulans gene regulatory protein AreA causes a conformational change at the DNA-binding site. Protein Expression Purif. 39(2): 152–159. https://doi.org/10.1016/j.pep.2004.10.017
- Zhao, D. and Huang, Z. (2016). Effect of His-Tag on Expression, Purification, and Structure of Zinc Finger Protein, ZNF191(243-368). Bioinorg Chem Appl. 2016: 1–6. https://doi.org/10.1155/2016/8206854
- Wang, B., Alam, S. L., Meyer, H. H., Payne, M., Stemmler, T. L., Davis, D. R. and Sundquist, W. I. (2003). Structure and Ubiquitin Interactions of the Conserved Zinc Finger Domain of Npl4. J Biol Chem. 278(22): 20225–20234. https://doi.org/10.1074/jbc.m300459200
- Schrader, N., Koerner, C., Koessmeier, K., Bangert, J. A., Wittinghofer, A., Stoll, R. and Vetter, I. R. (2008). The Crystal Structure of the Ran-Nup153ZnF2 Complex: a General Ran Docking Site at the Nuclear Pore Complex. Structure. 16(7): 1116–1125. https://doi.org/10.1016/j.str.2008.03.014
- Crotty, J. W., Etzkorn, C., Barbas, C. F., Segal, D. J. and Horton, N. C. (2005). Crystallization and preliminary X-ray crystallographic analysis of Aart, a designed six-finger zinc-finger peptide, bound to DNA. Acta Crystallogr Sect F Struct Biol Cryst Commun. 61(6): 573–576. https://doi.org/10.1107/s1744309105014363
- Zhang, L., Spratt, S. K., Liu, Q., Johnstone, B., Qi, H., Raschke, E. E., Jamieson, A. C., Rebar, E. J., Wolffe, A. P., Case, C. C., et al. (2000). Synthetic Zinc Finger Transcription Factor Action at an Endogenous Chromosomal Site. J Biol Chem. 275(43): 33850–33860. https://doi.org/10.1074/jbc.m005341200
- Ko, C. C., Ostermeier, M. and Lin, S. C. (2018). Dual column approach for the purification of zinc finger proteins by immobilized metal affinity chromatography. Process Biochem. 73: 204–210. https://doi.org/10.1016/j.procbio.2018.08.025
- Zhou, Z., Xiang, Y., Tong, A. and Lu, Y. (2014). Simple and Efficient Method to Purify DNA–Protein Conjugates and Its Sensing Applications. Anal Chem. 86(8): 3869–3875. https://doi.org/10.1021/ac4040554
- Liang, J., Azubel, M., Wang, G., Nie, Y., Kornberg, R. D., Beel, A. J. and Mattei, P. J. (2025). A universal method for the purification of C2H2 zinc finger arrays. PLoS One. 20(2): e0318295. https://doi.org/10.1371/journal.pone.0318295
- Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 72: 248–254. https://doi.org/10.1016/0003-2697(76)90527-3
- Liang, J., Azubel, M., Wang, G., Nie, Y., Kornberg, R. D., Beel, A. J. and Mattei, P. J. (2025). A universal method for the purification of C2H2 zinc finger arrays. PLoS One. 20(2): e0318295. https://doi.org/10.1371/journal.pone.0318295
Article Information
Publication history
Received: May 12, 2025
Accepted: Jun 27, 2025
Available online: Aug 1, 2025
Published: Aug 20, 2025
Copyright
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Liang, J., Azubel, M., Wang, G., Nie, Y., Kornberg, R. D., Beel, A. J. and Matteï, P. J. (2025). Production of Homogeneous, Functional Zinc-Finger Arrays in High Yield With Two Chromatographic Steps. Bio-protocol 15(16): e5420. DOI: 10.21769/BioProtoc.5420.
Category
Biochemistry > Protein > Isolation and purification
Biochemistry > Protein > Expression
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








