- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Silencing Arbuscular Mycorrhizal Fungal Gene Using Chitosan Nanoparticle-Mediated dsRNA Delivery System
Published: Vol 15, Iss 11, Jun 5, 2025 DOI: 10.21769/BioProtoc.5326 Views: 2645
Reviewed by: Shweta PanchalAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

In Vitro Hyphal Branching Assay Using Rhizophagus irregularis
Takaya Tominaga and Hironori Kaminaka
Aug 20, 2024 2338 Views

In Vitro Screening of Microbial Extracts Against the Oomycetes Phytophthora capsici and Pythium ultimum
Mónica Trigal Martínez [...] María Ángeles Vinuesa Navarro
Sep 20, 2025 1265 Views

A Reliable In Planta Inoculation and Antifungal Screening Protocol for Rhizoctonia solani-Induced Sheath Blight in Rice
Alinaj Yasin [...] Palash Deb Nath
Nov 5, 2025 1582 Views
Abstract
It has been discovered that many phytopathogenic fungi can absorb exogenous double-stranded RNAs (dsRNAs) to silence target genes, inhibiting fungal growth and pathogenicity for plant protection. In our recent report, the beneficial arbuscular mycorrhizal (AM) fungi are capable of acquiring external naked dsRNAs; however, whether the dsRNAs can be delivered into AM fungi through nanocarriers remains to be investigated. Here, we introduce a simple and advanced method for in vitro synthesizing chitosan (CS)/dsRNA polyplex nanoparticles (PNs) to silence the target gene in the AM fungus Rhizophagus irregularis. This method is straightforward, requiring minimal modifications, and is both efficient and eco-friendly, offering potential for rapid application in elucidating gene functions in AM fungi.
Key features
• The chitosan can carry the dsRNA derived from the AM fungus Rhizophagus irregularis.
• CS/dsRNA polyplex nanoparticles (PNs) can successfully silence the target gene in the AM fungus R. irregularis.
• CS/dsRNA PNs can be applied to the characterization of AM fungal genes via the spray-induced gene silencing (SIGS) approach.
• This protocol can be applied in asymbiotic and symbiotic cultures of AM fungi.
Keywords: Arbuscular mycorrhizal fungiGraphical overview
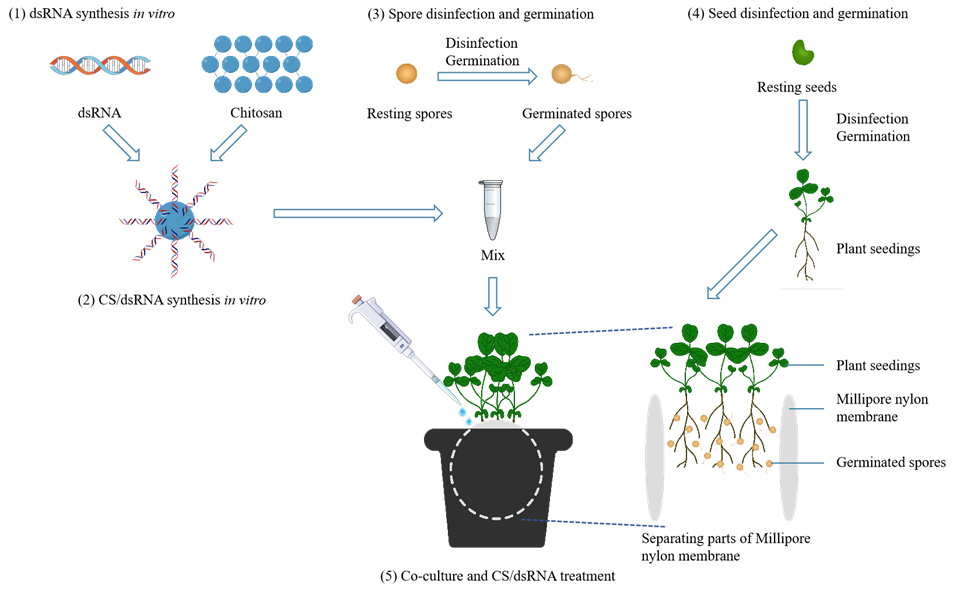
Overview of the chitosan/dsRNA gene silencing procedures
Background
Arbuscular mycorrhizal (AM) fungi are widely distributed in terrestrial ecosystems and can form symbiotic associations with 72% of land plants [1–3]. However, AM fungi are obligate symbionts that are not capable of completing their natural life cycle without the host plants [4]. Therefore, it is very difficult to characterize AM fungal gene functions by mutation methods such as gene knockout. RNA interference–based methods, such as HIGS (host-induced gene silencing) and VIGS (virus-induced gene silencing), can provide alternative tools for AM fungal gene knockdown during AM symbiosis [5–8]. Despite their potential, these host plant–dependent cross-kingdom RNAi methods have their limitations: HIGS is hindered by the host plant's transformation efficiency, while VIGS faces constraints due to the transient nature of gene silencing. Moreover, HIGS and VIGS approaches are not suitable for elucidating gene functions in AM fungi during the asymbiotic and presymbiotic stages.
In recent studies, some phytopathogenic fungi have been found to have a robust capacity to absorb exogenous double-stranded RNAs (dsRNAs) and small RNAs (sRNAs), which can silence target genes in fungi via the RNAi pathway, thereby reducing fungal growth, development, and pathogenicity [9–11]. The spray-induced gene silencing (SIGS) is an innovative and eco-friendly technology where application of gene-targeting dsRNAs to fungal materials can enable gene functional analysis in AM fungi [12]. However, SIGS applications remain limited because of the instability of RNA, which can be easily degraded when exposed to various environmental conditions. Also, continuous application of dsRNAs is required to maintain the RNAi effect [13–14]. Qiao et al. [15] described the use of artificial nanovesicles for dsRNA delivery in SIGS for crop protection. The results of this work inspired how nanocarriers can be used as new carriers to overcome RNA instability in SIGS for AM fungal gene analysis.
Chitosan (CS) is a natural polysaccharide derived from marine organisms with excellent biocompatibility and biodegradability [16]. The interaction between the positively charged amino groups in chitosan molecules can form stable complexes with the negatively charged phosphate groups in RNA chains. Therefore, chitosan may be an ideal delivery carrier for dsRNAs, sRNAs, and other molecules [11,16,17]. Importantly, the use of nanoparticles as a carrier for dsRNA not only protects dsRNAs and slows down their degradation rate but also effectively delivers dsRNAs into the cells of living organisms to achieve precise gene silencing.
Here, we synthesize chitosan/dsRNA polyplex nanoparticles (PNs) that target the RiABCG6.3 gene (Gene ID: XM_025322239.1) in Rhizophagus irregularis. RiABCG6.3 is a homolog of the known ABCG-1 gene, MoCDR1, in rice-blast fungus Magnaporthe oryzae, which facilitates appressorium formation [18]. We can speculate the role of RiABCG6.3 in appressorium or hyphopodium formation in R. irregularis and use chitosan/dsRNA PNs to silence the transcripts of RiABCG6.3 to explore its function at the early stages of AM symbiosis. By introducing this chitosan/dsRNA PNs complex to R. irregularis and mycorrhizal roots of Astragalus sinicus seedlings, the expression of RiABCG6.3 is successfully silenced. This approach is straightforward, requiring minimal modifications, and is both efficient and eco-friendly. Overall, the applications of chitosan/dsRNA PNs to fungal materials can elucidate gene functions in AM fungi.
In this protocol, we outline the detailed procedures for the in vitro synthesis and characterization of chitosan/dsRNA PNs (Graphical overview) and assessment of the efficiency of gene silencing via RT-qPCR assay.
Materials and reagents
Biological materials
1. AM fungal spores [Rhizophagus irregularis DAOM 197198 spores’ solutions with 10 mM sodium citrate buffer (pH 6.0) were kindly provided by Dr. Nianwu Tang, Guangxi Institute of Botany, Guilin, China].
2. Astragalus sinicus seeds (obtained from the National Key Laboratory of Agricultural Microbiology at Huazhong Agricultural University, Wuhan, China)
Reagents
1. Nuclease-free water (Sigma-Aldrich, catalog number: 3098)
2. Chitosan (Sigma-Aldrich, catalog number: C3646)
3. Na2SO4 (Sigma-Aldrich, catalog number: 239313)
4. Acetic acid (Sigma-Aldrich, catalog number: A6283)
5. Sodium acetate (Sigma-Aldrich, catalog number: 229873)
6. Micrococcal nuclease enzyme (NEB, catalog number: M0247S)
7. SDS (Sigma-Aldrich, catalog number: 75746)
8. Fluorescein-12-UTP (Sigma-Aldrich, catalog number: 11427857910)
9. Gel Recovery kit (OMEGA Bio-Tek, catalog number: D2500)
10. HiScribe® T7 High Yield RNA Synthesis kit (NEB, catalog number: E2040L)
11. RNase-free DNase I (Sigma-Aldrich, catalog number: 69182)
12. GR24 (Strigolactone analog) (Macklin, catalog number: S885104)
13. Chloramine T (Sigma-Aldrich, catalog number: 402869)
14. Streptomycin (Sigma-Aldrich, catalog number: 85886)
15. Gentamycin (Sigma-Aldrich, catalog number: 345814)
16. Agarose (Sigma-Aldrich, catalog number: A9539)
17. WGA488 (Alexa Fluor 488 labeled wheat germ agglutinin) (Thermo Fisher Scientific, catalog number: W849)
18. DNA marker (Takara, catalog number: 3427Q)
19. RNA marker (Takara, catalog number: TCH023)
20. 10× DNA loading buffer (Vazyme, catalog number: P022-01)
21. RNA loading buffer (Sangon, catalog number: B548317)
22. NaAc (Sigma-Aldrich, catalog number: S7670)
Solutions
1. Sodium acetate buffer, pH 4.5 (see Recipes)
2. 2.5 mol/L Na2SO4 (see Recipes)
3. 0.02% chitosan working solution (see Recipes)
Recipes
1. Sodium acetate buffer, pH 4.5
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| NaAc | 0.1 M | 18 g |
| Acetic acid | 0.1 M | 9.8 mL |
| Nuclease-free water | n/a | up to 1,000 mL |
| Total (optional) | n/a | 1,000 mL |
2. 2.5 mol/L Na2SO4
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Na2SO4 | 2.5 M | 17.755 g |
| Nuclease-free water | n/a | up to 50 mL |
| Total (optional) | n/a | 50 mL |
3. 0.02% chitosan working solution
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Chitosan | 0.02% | 0.02 g |
| Nuclease-free water | n/a | up to 100 mL |
| Total (optional) | n/a | 100 mL |
Laboratory supplies
1. Millipore nylon filter membrane (JINTENG, catalog number: LG05-112)
2. Quartz sand (Macklin, catalog number: S861671)
3. Nuclease-free centrifuge tubes (Sangon Biotech, catalog number: F601621-0010; F607620-0005)
4. Pipette tips (Vazyme, catalog number: PTB00122, PTB02022, PTB10022)
5. Microscope slides (Sangon, catalog number: F518111-0001) and coverslips (Sangon, catalog number: F518117-0001)
6. Plastic pots (6 × 6 cm diameter) (Hongyue, catalog number: 26541)
7. Sterile Petri dishes (90 mm diameter) (Huankai, catalog number: 88574540)
8. Fine tip tweezers (Sangon, catalog number: F519231)
9. Spray bottles (Hongyue, catalog number: 30700)
10. Personal cell phone (in our case, iPhone 13)
Equipment
1. Thermal cycler CFX96 touch real-time PCR detection system (Bio-Rad, CFX ConnectTM, catalog number: 1855200)
2. Thermal cycler PCR (Bio-Rad, catalog number: S1000)
3. Analytical balance (Mettler Toledo, model: Tle204/02)
4. Vortex mixer (Thermo Fisher Scientific, catalog number: 88882012)
5. pH meter (METTLER TOLEDO, catalog number: 30254104)
6. Gel image analysis system (Bio-Rad, model: T2A)
7. Electrophoresis power supply (Liuyi Biotechnology, model: DYY-)
8. Electrophoresis chamber (Liuyi Biotechnology, catalog number: 122-3146)
9. Confocal microscope (Zeiss, model: LSM 710)
10. Sceintz-48 high-throughput tissue grinder (Sceintz, model: 18-G009)
11. Fluorescence microscope (Nikon, catalog number: Y-TV55)
12. Stereo fluorescence microscope (Nikon, catalog number: SMZ18)
13. Nucleic acid protein analyzer NanoDrop 2000 (Thermo Fisher Scientific, catalog number: ND-2000)
14. Plant growth chamber
15. Thermostatic water bath (OLT, catalog number: HH-24)
Software and datasets
1. BLASTn and BLASTp (http://blast.ncbi.nlm.nih.gov/Blast.cgi)
2. Mega11.0 (https://www.megasoftware.net/dload_win_beta)
3. siRNA-Finder (si-Fi) software [19] (https://github.com/snowformatics/siFi21-)
4. GraphPad Prism v9.0.0 (https://www.graphpad.com/scientific-software/prism/)
5. Integrated DNA Technologies (https://sg.idtdna.com/sessionTimeout.aspx/)
Procedure
A. AM fungus and plant growth conditions
1. Store R. irregularis DAOM 197198 spore solutions with 10 mM sodium citrate buffer (pH 6.0) at 4 °C until use.
2. Perform surface sterilization of spores as reported previously [7]. For preparing the germinating spores, place them in sterile water containing 10-7 M GR24 and germinate at 25 °C in darkness for 7 days.
3. Before germination, soak A. sinicus seeds in 75% alcohol for 10 min, wash with sterile water three times, surface sterilize in 3% NaClO for 20 min, wash with sterile water 3–4 times, and finally immerse seeds in sterile water in a 28 °C incubator overnight. Sow surface-sterilized seeds on 0.6% agar plates.
4. After 7 days, transfer A. sinicus seedlings to pots containing 400 g of sterile quartz sand inoculated with 400 germinating spores (see General notes). Then, place the A. sinicus seedlings with R. irregularis into the conditional chamber (cycles of 16 h light at 25 °C and 8 h dark at 18 °C) for plant growth.
B. dsRNA design and synthesis in vitro
1. Download the CDS sequences of target genes from AM fungus R. irregularis from the Genbank database and use the siRNA-Finder (si-Fi) software [19] to predict sequences with likely efficient RNAi of the target genes identified in R. irregularis. Select sequences for dsRNA syntheses at about 200–300 bp and use NCBI BLASTn tool to confirm that none of the sequences were conserved in the host plant (A. sinicus) or other eukaryotic organisms.
2. Use the PrimerQuestTM tool from the webpage of Integrated DNA Technologies to design gene-specific primers with a length of about 20–25 bp and a specific amplification length of about 200–300 bp. Add T7 promoter sequences at the 5' end of the forward and reverse primers and submit to Sangon Biotech (Guangzhou, China) (https://www.sangon.com/) for primer synthesis.
3. Amplify the target gene and GFP gene products from mycorrhizal roots and GFP-containing pUG36 plasmid [20], respectively, using gene-specific primers. Use the Gel Recovery kit for gel recovery and purification of PCR products. Detect the concentration and purity of the recovered products by NanoDrop 2000. Synthesize dsRNA in vitro using the HiScribe® T7 High Yield RNA Synthesis kit according to the manufacturer’s instructions and using GFP as a control. Assemble the reactions as described in Table 1.
Table 1. Reactions for in vitro dsRNA synthesis
| Reagent | Volume | Final concentration or volume |
|---|---|---|
| 10× reaction buffer | 1.5 μL | |
| ATP (100 mM) | 1.5 μL | 7.5 mM |
| GTP (100 mM) | 1.5 μL | 7.5 mM |
| CTP (100 mM) | 1.5 μL | 7.5 mM |
| UTP (100 mM) | 1.5 μL | 7.5 mM |
| Template DNA | 1 μg | |
| T7 RNA polymerase mix | 1.5 μL | |
| Nuclease-free water | Up to 20 μL |
4. Remove template DNA using RNase-free DNase I and inactivate at 65 °C for 10 min. Then, measure the dsRNA concentration and purity using NanoDrop 2000 as mentioned above.
5. Take 0.5–1 μL of the synthesized dsRNA to detect its integrity by agarose gel electrophoresis (Figure 1).
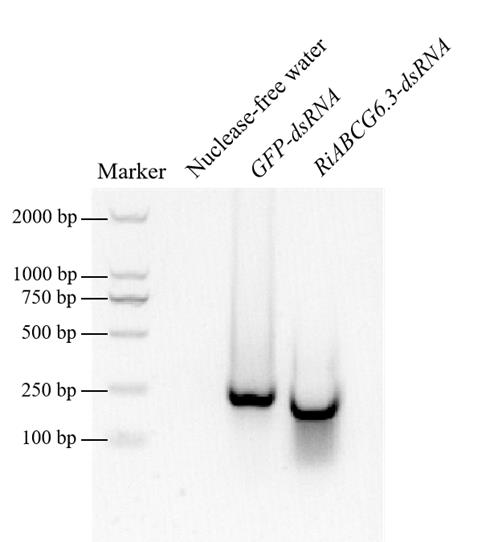
Figure 1. In vitro synthesized dsRNA. In vitro synthesis of GFP-dsRNA and RiABCG6.3-dsRNA, with nuclease-free water used as a control.
C. Chitosan-dsRNA PNs preparation and selection of optimal fusion ratio
1. Prepare the sodium acetate buffer (see Recipes).
2. Dissolve chitosan in sodium acetate buffer to formulate a 0.02% working solution for backup.
3. Pipette different concentrations of dsRNA into clean centrifuge tubes separately, add 2 μL of 2.5 mol/L Na2SO4 to the centrifuge tubes, and make up to a total of 100 μL with nuclease-free water. Mix gently using a pipette and then centrifuge briefly at 100× g for 30 s. After sufficient mixing, add 100 μL of 0.02% chitosan working solution. The final ratios of dsRNA to chitosan are 5:1, 4:1, 3:1, 2:1, 1:1, and 1:2.
4. Let the centrifuge tubes stand at room temperature for 2 min and then place them in a 55 °C water bath for 1 min before rapidly vortexing for 30 s.
5. Leave them to stand for 10 min; during this time, the dsRNA is absorbed on the surface of chitosan to form stable chitosan/dsRNA nanoparticles.
6. Detect 10 μL of CS/dsRNA nanoparticle solutions by agarose gel electrophoresis (agarose concentration 1%) (Figure 2) and select the optimal fusion ratio according to the electrophoresis results. As shown in Figure 2, when the ratios of dsRNA to chitosan are 1:1 and 1:2, chitosan can load dsRNA completely; when the ratio is 2:1, there is a small amount of dsRNA that is not loaded, forming diffuse bands.
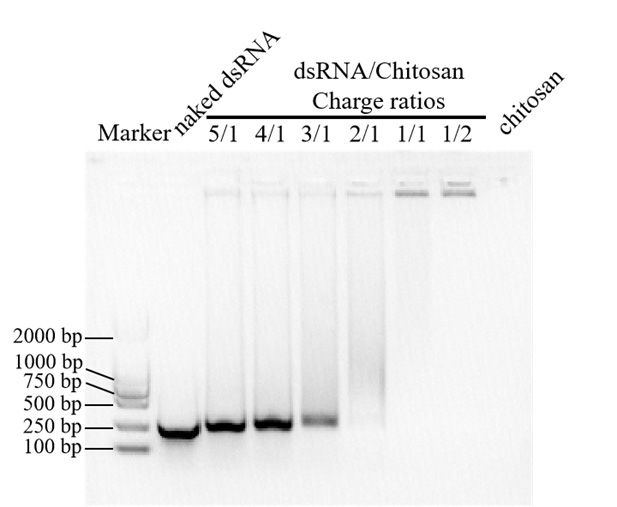
Figure 2. Chitosan was effectively loaded with RiABCG6.3-dsRNA to help protect against degradation. Gel electrophoresis assay showing dsRNA retardation by chitosan at various dsRNA/chitosan mass ratios (5:1, 4:1, 3:1, 2:1, 1:1, and 1:2).
D. Chitosan-dsRNA PNs stability analysis
1. Add 1 μL of micrococcal nuclease that was diluted tenfold to the chitosan-dsRNA PNs (1:1 ratio of dsRNA:chitosan) for 10–90 min at 37 °C to assess stability. Include a naked dsRNA treatment as a negative control. Before starting, measure dsRNA concentrations on a NanoDrop 2000.
2. Add 0.2% SDS solution to each solution at 25 °C for 10 min, releasing the dsRNA from chitosan nanoparticles.
3. Observe degradation by nucleic acid gel electrophoresis (Figure 3).
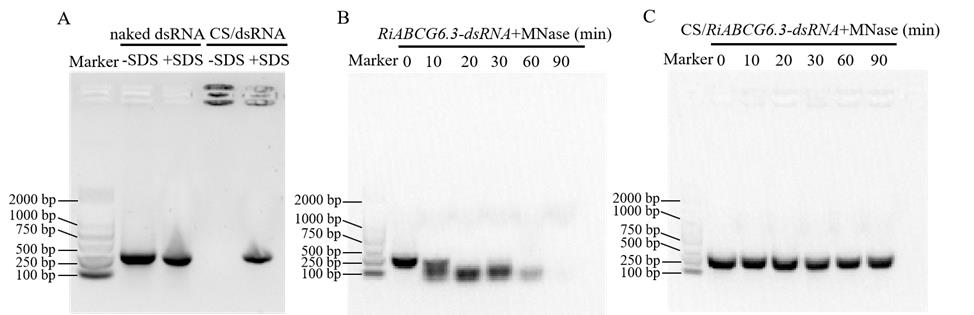
Figure 3. Enhanced stability of chitosan (CS)/dsRNA. Exogenous nuclease protection of 200 ng dsRNA and (A) protection of dsRNA from natural degradation in chitosan. (B) Using naked dsRNA as a control, (C) formulation with chitosan prevents migration into the gel and binding of nucleic acid dye. After binding to chitosan, dsRNA can be released by the addition of 0.2% SDS.
E. Treatment of R. irregularis–A. sinicus co-culture with chitosan-dsRNAs
1. To prepare germinating spores, place them in sterile water containing 10-7 M GR24 and an equal volume of 400 ng/μL of CS/RiABCG6.3-dsRNA. Use sterile water, CS/GFP-dsRNA, GFP-dsRNA, and RiABCG6.3-dsRNA as controls. Germinate spores at 25 °C in the dark for 7 days.
2. Transplant A. sinicus seedlings into a microporous filter membrane in the center of the pots with quartz sand: transplant 3 seedlings per pot and inoculate each pot with ~500 germinating spores of R. irregularis (see General notes).
3. After 3 days of incubation, using a pipette, spray 300 μL of 200 ng/μL of CS/RiABCG6.3-dsRNA onto the root surfaces inoculated with R. irregularis spores in each pot to enhance treatment. Use sterile water, CS/GFP-dsRNA, GFP-dsRNA, and RiABCG6.3-dsRNA as controls and set up 18 replicates for each treatment. Every 7 days, randomly select 6–8 replicates to observe AM fungal colonization. Figure 4 shows plant growth phenotypes after 21 days of different treatments.
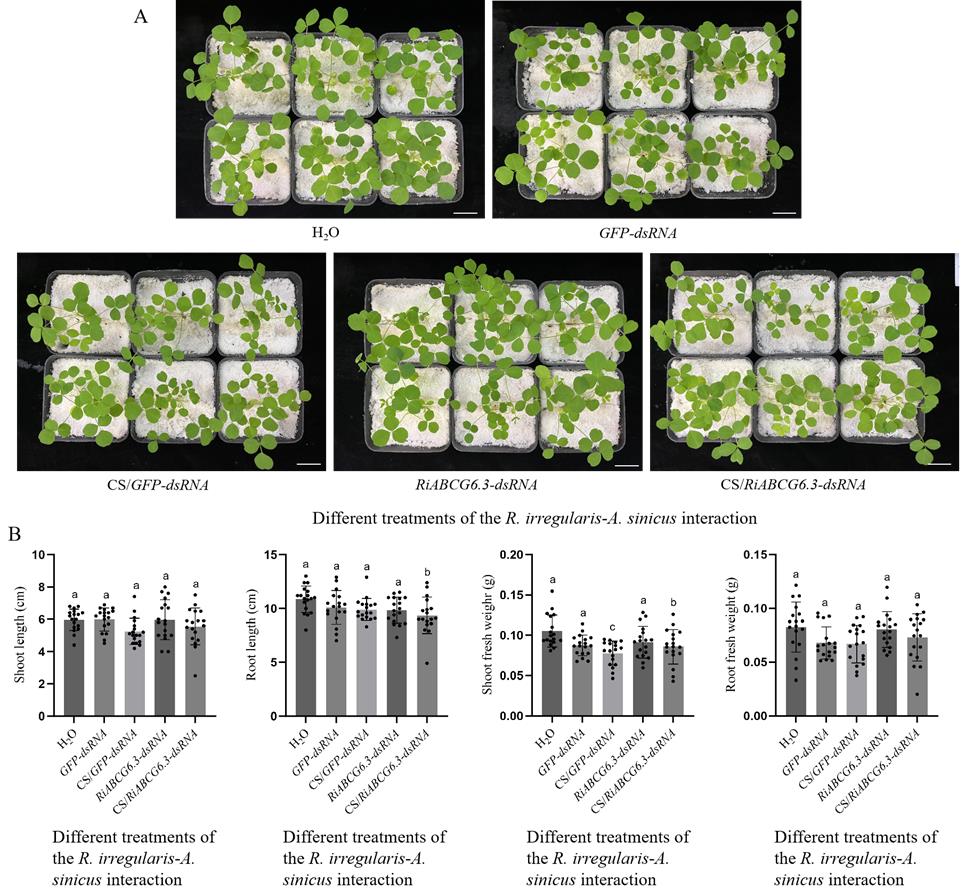
Figure 4. Different treatments were applied to the R. irregularis–A. sinicus interaction. (A) Growth performances of 21-day-old A. sinicus seedlings 21 days post inoculation. Scale bars, 2 cm. (B) Plant growth index of A. sinicus seedlings under different treatments. All values are means ± SD; different lowercase letters indicate significant differences (n = 18, P < 0.05, Duncan’s test).
4. Collect mycorrhizal roots of A. sinicus and stain one part of the root samples with 5 μg/mL WGA488 to detect colonization levels (see data analysis) [12,21]. Use the other part of the roots for total RNA extraction to detect the silencing efficiency of RiABCG6.3 (Figure 5); the R. irregularis RiTRK1 gene in the cAMP-PKA pathway is used as a marker gene for hyphopodia formation [22].
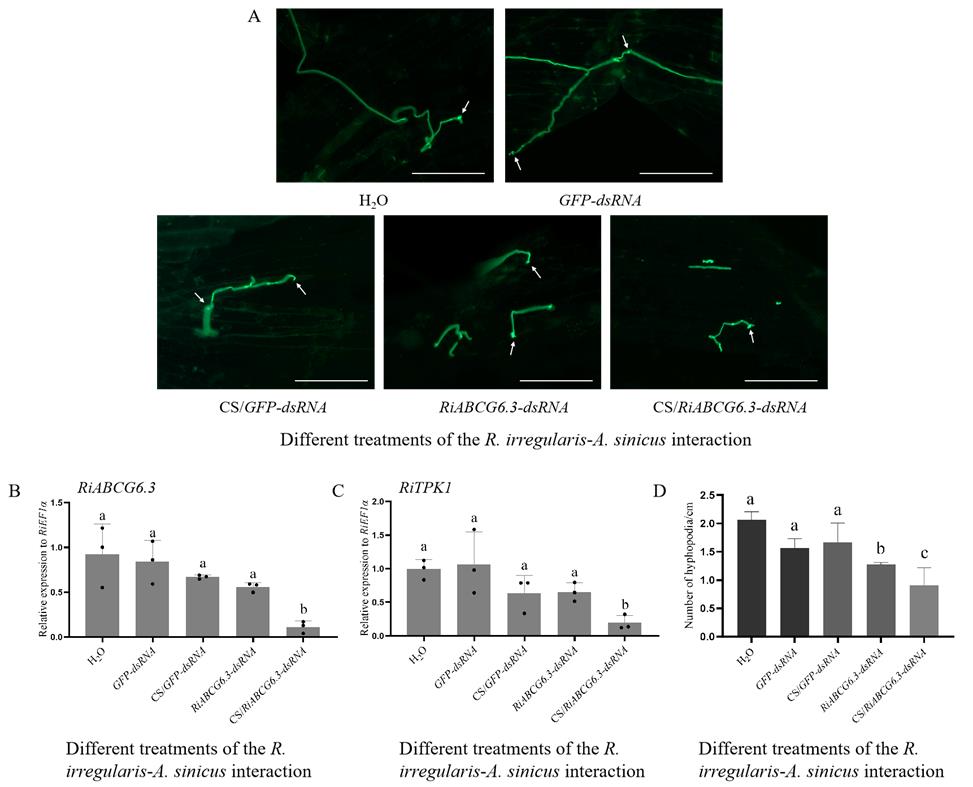
Figure 5. RiABCG6.3-dsRNA-mediated gene silencing. (A) Confocal microscopy images of hyphopodia of R. irregularis–A. sinicus interaction after 21 days of chitosan (CS)/dsRNA PNs and control treatments. The arrows in panel A indicate hyphopodia. Scale bars, 100 μm. (B–D) Relative expression of RiABCG6.3 (B) and RiTPK1 (C), and hyphopodia on the A. sinicus roots with R. irregularis exposed to different dsRNAs or CS/dsRNAs applications for 21 days (D). R. irregularis RiEF1α was used as the reference gene for expression normalization, while RiTRK1 served as a marker gene for hyphopodia formation. The values are means ± SD; different lowercase letters indicate significant differences (n = 3, P < 0.05, Duncan’s test).
Data analysis
1. For all experiments, include at least three biological replicates.
2. To calculate the number of hyphopodia, refer to previous methods as described by Breuninger & Requena and Fan et al. [12,21]. The roots are digested with 10% (w/v) KOH solution at 37 °C for 30 min, after which the KOH solution is neutralized in 2% (v/v) hydrochloric acid. Roots are washed at least three times with sterile distilled water, stained with 5 μg/mL WGA488, cut into approximately 1 cm in length, arranged on a slide in a 4 × 5 grid pattern, and then observed under a fluorescence microscope to calculate the number of hyphopodia per centimeter.
3. For the RT-qPCR assay, include at least three biological and technical replicates.
4. GraphPad Prism v9.0.0 was used for graphing and statistical analyses. Different letters on the graphs indicate significant differences among phenotypes or treatments. All data were presented as averages ± SD or ± SE. One-way ANOVA with Tukey's test was used for multiple comparison analysis. A value of P < 0.05 was considered to be statistically significant.
Validation of protocol
Chitosan can carry dsRNA to form PNs (Figures 2 and 3). Then, CS/RiABCG6.3 PNs can successfully silence the RiABCG6.3 gene from R. irregularis and have a better silencing effect than naked RiABCG6.3-dsRNA (Figures 4 and 5), which sufficiently validates this protocol.
General notes and troubleshooting
General notes
1. For the surface sterilization of spores, spores were soaked in 2% chloramine T for 15 min, washed at least three times in sterile water, and immersed in a mixture of antibiotic solution containing 200 mg/L streptomycin and 100 mg/L gentamycin for 10 min.
2. The method is also suitable for other AM fungal species, but it may be necessary to modify some experimental conditions, such as the ratio of dsRNA to chitosan.
3. The length of the synthesized target gene fragment is recommended to be about 200–300 bp.
4. A schematic diagram of the use of two Millipore nylon membranes (50 mm in diameter, pore diameter 0.8 μm) to assist R. irregularis–A. sinicus interaction is shown in graphical overview.
5. For step 5 of the Graphical overview (“Co-culture and CS/dsRNA treatment”), in short, select three uniform A. sinicus seedlings and place them on a Millipore nylon membrane. Then, using a pipette, transfer 300 μL of 200 ng/μL CS/dsRNA solution containing 400 R. irregularis spores and spray it onto the root surfaces of A. sinicus. Subsequently, A. sinicus seedlings are covered with another nylon membrane. Vertically insert the entire assembly into the center of a square pot containing quartz sand, ensuring that one-fifth of the nylon membrane remains exposed above the quartz sand.
6. Synthesized nanoparticles can be stored at -20 °C for a short period of 1–2 weeks, avoiding repeated freezing and thawing. The integrity of dsRNA should be detected by agarose gel electrophoresis before use.
Troubleshooting
Problem 1: When synthesizing dsRNA, the detected strips are dragging or diffusing.
Possible cause: The dsRNA has degraded.
Solution: Synthesize dsRNA in a clean and nuclease-free environment, using nuclease-free reagents and water, and ensure that the template used for synthesis is sufficiently fresh.
Problem 2: When synthesizing chitosan (CS)/dsRNA PNs, chitosan cannot carry dsRNA effectively.
Possible cause: There may be a problem with the preparation of the chitosan solution, or dsRNA has been stored for too long.
Solution: Prepare the CS working solution again or resynthesize the dsRNA. Use nuclease-free water and pipette tips. The reagents (such as Na2SO4 solution) should be filtered to remove bacteria. Avoid using chitosan solutions that have been stored at room temperature for too long for CS/dsRNA PNs preparation.
Acknowledgments
This work was financially supported by the National Natural Science Foundation of China (32370108 and 32170116) and the State Key Laboratory of Nutrient Use and Management (KF2024-4).
Competing interests
Authors declare no competing interests.
References
- Genre, A., Lanfranco, L., Perotto, S. and Bonfante, P. (2020). Unique and common traits in mycorrhizal symbioses. Nat Rev Microbiol. 18(11): 649–660. https://doi.org/10.1038/s41579-020-0402-3
- Rich, M. K., Vigneron, N., Libourel, C., Keller, J., Xue, L., Hajheidari, M., Radhakrishnan, G. V., Le Ru, A., Diop, S. I., Potente, G., et al. (2021). Lipid exchanges drove the evolution of mutualism during plant terrestrialization. Science. 372(6544): 864–868. https://doi.org/10.1126/science.abg0929
- Duan, S., Feng, G., Limpens, E., Bonfante, P., Xie, X. and Zhang, L. (2024). Cross-kingdom nutrient exchange in the plant–arbuscular mycorrhizal fungus–bacterium continuum. Nat Rev Microbiol. 22(12): 773–790. https://doi.org/10.1038/s41579-024-01073-7
- Sugiura, Y., Akiyama, R., Tanaka, S., Yano, K., Kameoka, H., Marui, S., Saito, M., Kawaguchi, M., Akiyama, K., Saito, K., et al. (2020). Myristate can be used as a carbon and energy source for the asymbiotic growth of arbuscular mycorrhizal fungi. Proc Natl Acad Sci USA. 117(41): 25779–25788. https://doi.org/10.1073/pnas.2006948117
- Helber, N., Wippel, K., Sauer, N., Schaarschmidt, S., Hause, B. and Requena, N. (2011). A Versatile Monosaccharide Transporter That Operates in the Arbuscular Mycorrhizal Fungus Glomus sp Is Crucial for the Symbiotic Relationship with Plants. Plant Cell. 23(10): 3812–3823. https://doi.org/10.1105/tpc.111.089813
- Kikuchi, Y., Hijikata, N., Ohtomo, R., Handa, Y., Kawaguchi, M., Saito, K., Masuta, C. and Ezawa, T. (2016). Aquaporin‐mediated long‐distance polyphosphate translocation directed towards the host in arbuscular mycorrhizal symbiosis: application of virus‐induced gene silencing. New Phytol. 211(4): 1202–1208. https://doi.org/10.1111/nph.14016
- Xie, X., Lin, H., Peng, X., Xu, C., Sun, Z., Jiang, K., Huang, A., Wu, X., Tang, N., Salvioli, A., et al. (2016). Arbuscular Mycorrhizal Symbiosis Requires a Phosphate Transceptor in the Gigaspora margarita Fungal Symbiont. Mol Plant. 9(12): 1583–1608. https://doi.org/10.1016/j.molp.2016.08.011
- Xie, X., Lai, W., Che, X., Wang, S., Ren, Y., Hu, W., Chen, H. and Tang, M. (2022). A SPX domain‐containing phosphate transporter from Rhizophagus irregularis handles phosphate homeostasis at symbiotic interface of arbuscular mycorrhizas. New Phytol. 234(2): 650–671. https://doi.org/10.1111/nph.17973
- Wang, M., Weiberg, A., Lin, F. M., Thomma, B. P. H. J., Huang, H. D. and Jin, H. (2016). Bidirectional cross-kingdom RNAi and fungal uptake of external RNAs confer plant protection. Nat Plants. 2(10): e151. https://doi.org/10.1038/nplants.2016.151
- Qiao, L., Lan, C., Capriotti, L., Ah-Fong, A., Sanchez, J. N., Hamby, R., Heller, J., Zhao, H., Glass, N. L., Judelson, H. S., et al. (2021). Spray-induced gene silencing for disease control is dependent on the efficiency of pathogen RNA uptake. Plant Biotechnol J. 19(9):1756–1768. https://doi.org/10.1101/2021.02.01.429265
- Ray, P., Sahu, D., Aminedi, R. and Chandran, D. (2022). Concepts and considerations for enhancing RNAi efficiency in phytopathogenic fungi for RNAi-based crop protection using nanocarrier-mediated dsRNA delivery systems. Front Fungal Biol. 3: e977502. https://doi.org/10.3389/ffunb.2022.977502
- Fan, X., Zhou, X., He, J., Xie, H., Tang, N., Tang, M. and Xie, X. (2025). Spray‐induced gene silencing of three G‐protein signaling genes from the arbuscular mycorrhizal fungus Rhizophagus irregularis inhibits spore germination and hyphopodium formation. New Phytol. e70091. https://doi.org/10.1111/nph.70091
- Mitter, N., Worrall, E. A., Robinson, K. E., Xu, Z. P. and Carroll, B. J. (2017). Induction of virus resistance by exogenous application of double-stranded RNA. Curr Opin Virol. 26: 49–55. https://doi.org/10.1016/j.coviro.2017.07.009
- Abdellatef, E., Kamal, N. M. and Tsujimoto, H. (2021). Tuning Beforehand: A Foresight on RNA Interference (RNAi) and In Vitro-Derived dsRNAs to Enhance Crop Resilience to Biotic and Abiotic Stresses. Int J Mol Sci. 22(14): 7687. https://doi.org/10.3390/ijms22147687
- Qiao, L., Niño‐Sánchez, J., Hamby, R., Capriotti, L., Chen, A., Mezzetti, B. and Jin, H. (2023). Artificial nanovesicles for dsRNA delivery in spray‐induced gene silencing for crop protection. Plant Biotechnol J. 21(4): 854–865. https://doi.org/10.1111/pbi.14001
- Zhou, H., Wan, F., Jian, Y., Guo, F., Zhang, M., Shi, S., Yang, L., Li, S., Liu, Y., Ding, W., et al. (2023). Chitosan/dsRNA polyplex nanoparticles advance environmental RNA interference efficiency through activating clathrin-dependent endocytosis. Int J Biol Macromol. 253: 127021. https://doi.org/10.1016/j.ijbiomac.2023.127021
- Yan, S., Ren, B. and Shen, J. (2020). Nanoparticle‐mediated double‐stranded RNA delivery system: A promising approach for sustainable pest management. Insect Sci. 28(1): 21–34. https://doi.org/10.1111/1744-7917.12822
- Wang, J., Xiao, C., Liang, S., Noman, M., Cai, Y., Zhang, Z., Zhu, X., Chai, R., Qiu, H., Hao, Z., et al. (2025). Comparative functional analysis of a new CDR1-like ABC transporter gene in multidrug resistance and virulence between Magnaporthe oryzae and Trichophyton mentagrophytes. Cell Commun Signaling. 23(1): 69. https://doi.org/10.1186/s12964-024-02022-w
- Lück, S., Kreszies, T., Strickert, M., Schweizer, P., Kuhlmann, M. and Douchkov, D. (2019). siRNA-Finder (si-Fi) Software for RNAi-Target Design and Off-Target Prediction. Front Plant Sci. 10: e01023. https://doi.org/10.3389/fpls.2019.01023
- Xie, X., Lai, W., Che, X., Wang, S., Ren, Y., Hu, W., Chen, H. and Tang, M. (2022). A SPX domain‐containing phosphate transporter from Rhizophagus irregularis handles phosphate homeostasis at symbiotic interface of arbuscular mycorrhizas. New Phytol. 234(2): 650–671. https://doi.org/10.1111/nph.17973
- Breuninger, M. and Requena, N. (2004). Recognition events in AM symbiosis: analysis of fungal gene expression at the early appressorium stage. Fungal Genet Biol. 41(8): 794–804. https://doi.org/10.1016/j.fgb.2004.04.002
- Zhou, X., Li, J., Tang, N., Xie, H., Fan, X., Chen, H., Tang, M. and Xie, X. (2021). Genome-Wide Analysis of Nutrient Signaling Pathways Conserved in Arbuscular Mycorrhizal Fungi. Microorganisms. 9(8): 1557. https://doi.org/10.3390/microorganisms9081557
Article Information
Publication history
Received: Feb 25, 2025
Accepted: Apr 24, 2025
Available online: May 16, 2025
Published: Jun 5, 2025
Copyright
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Yan, C., Wang, Y., Guo, Q., Huan, H., Wang, S., Fan, X. and Xie, X. (2025). Silencing Arbuscular Mycorrhizal Fungal Gene Using Chitosan Nanoparticle-Mediated dsRNA Delivery System. Bio-protocol 15(11): e5326. DOI: 10.21769/BioProtoc.5326.
Category
Microbiology > Microbe-host interactions > Fungus
Molecular Biology > Nanoparticle > Plan-derived nanoparticles
Plant Science > Plant immunity > Host-microbe interactions
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








