- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Antibody Purification Using Spy Chemistry
Published: Vol 15, Iss 8, Apr 20, 2025 DOI: 10.21769/BioProtoc.5286 Views: 1863
Reviewed by: Alessandro DidonnaDawid S ZylaCatherine Hurd

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
Analysis of Direct Interaction between Viral DNA-binding Proteins by Protein Pull-down Co-immunoprecipitation Assay
Ana Lechuga [...] Modesto Redrejo-Rodríguez
Jan 5, 2018 12399 Views
Visible Immunoprecipitation (VIP) Assay: a Simple and Versatile Method for Visual Detection of Protein-protein Interactions
Yohei Katoh [...] Kazuhisa Nakayama
Jan 5, 2018 13588 Views
Detection of Protein Interactions in the Cytoplasm and Periplasm of Escherichia coli by Förster Resonance Energy Transfer
Nils Y. Meiresonne [...] Tanneke den Blaauwen
Jan 20, 2018 14430 Views
Abstract
Antibody purification is a fundamental technology for therapeutic and diagnostic applications. While traditional methods like ammonium sulfate precipitation and polyethylene glycol precipitation are cost-effective, they often result in low purity and require multiple purification steps. Protein A–based affinity chromatography, the gold standard for antibody purification, provides high specificity but can be further improved to increase loading capacity and reduce costs. In this protocol, we introduce a novel approach for purifying high-quality, high-purity antibodies from complex samples using SpyFixer/Z domain–modified resin. This method utilizes Spy chemistry for site-specific immobilization of the Z domain of Protein A, significantly enhancing antibody loading capacity up to 200 mg/mL resin and ensuring stable, durable immobilization. Using this protocol, we achieved >90% purity of human immunoglobulin G (hIgG) from diverse sources, including E. coli cell lysates, human serum, and industrial fermentation broth. The SpyFixer/Z domain–modified resin protocol is simple, cost-effective, and offers a robust, scalable solution for efficient antibody purification in bioengineering applications. This immobilization scheme based on Spy chemistry can also be extended to other immunoglobulin-binding proteins, such as Protein G and Protein L, to develop high-efficiency affinity resins.
Key features
• This protocol builds upon purification methods developed by Lin’s lab [1], providing more detailed steps than the previously published study.
• The protocol offers a useful and standardized approach for purifying antibodies and Fc-fusion proteins.
• The SpyFixer/Z domain–modified resin is easy to prepare, reusable, and offers a cost-effective solution.
Keywords: Antibody purificationGraphical overview
Background
Antibodies are indispensable in modern medicine, playing a crucial role in therapeutic and diagnostic applications. The global monoclonal antibody (mAb) market has experienced rapid growth, projected to reach USD 263.28 billion in 2024 and USD 792.37 billion by 2032 [2]. To date, more than 150 recombinant antibody-based drugs have been approved by the FDA for the treatment of various diseases, such as cancer, rheumatic diseases, and infectious diseases [3]. As production cost pressures shift from fermentation to downstream processing, purification has become a significant bottleneck in antibody manufacturing.
Traditional purification methods such as ammonium sulfate precipitation and polyethylene glycol (PEG) precipitation are cost-effective but often result in co-precipitation with impurities, leading to insufficient purity and requiring multiple purification steps [4,5]. In contrast, Staphylococcal Protein A–based affinity chromatography has emerged as the gold standard for antibody purification, relying on the specific interaction between the immunoglobulin G (IgG) Fc region and immobilized Protein A ligands [6]. Most antibody fragments typically lack an Fc domain and do not bind to Protein A resins with high affinity [7]. Alternative immunoglobulin-binding proteins, such as Protein G (30 kDa) and Protein L (76–106 kDa), offer affinity-based purification strategies for antibody fragments, including antigen-binding fragments (Fabs), single-chain variable fragments (scFvs), and single-domain antibody fragments (dAbs), which typically lack an Fc domain and do not effectively bind to Protein A resins [8]. The use of Protein G involves low pH elution conditions and higher cost compared to Protein A resins [9,10]. Protein L binds specifically to antibodies containing certain kappa light chain subtypes, but this specificity limits its application in monoclonal antibody purification processes [6,9].
As an alternative affinity ligand, the Z domain—derived from the B domain of Protein A—has garnered attention for its advantages over full-length Protein A (~42 kDa), including its smaller size (~6.6 kDa), ease of engineering, higher stability, and comparable affinity [11,12]. These properties make the Z domain a promising candidate for antibody affinity purification. Additionally, the SpyTag/SpyCatcher system, a peptide-protein pair that spontaneously forms a covalently linked complex, has demonstrated significant potential for site-specific ligand immobilization [13]. To enhance immobilization efficiency and retention of binding activity, we employed SpyFixer, a variant of SpyCatcher, to site-specifically immobilize the Z domain onto a resin, generating SpyFixer/Z domain–modified resin [1].
Here, we develop an advanced antibody purification protocol using SpyFixer/Z domain–modified resin. This method achieves >90% purity and reusability, comparable to conventional Protein A–based methods while offering several advantages such as the use of inexpensive epoxy resins and increased antibody loading capacity [1]. These improvements provide a more cost-effective and scalable solution for antibody purification, with broad applications in biotechnology and pharmaceuticals. Using this protocol, we successfully purified IgG directly from complex samples, including E. coli cell lysates, human serum, and industrial fermentation broth [1].
Materials and reagents
Biological materials
1. E. coli BL21(DE3) competent cells (TransGen Biotech, catalog number: CD901-02)
2. Human immunoglobulin G (hIgG) (Sigma-Aldrich, catalog number: I4506)
Reagents
1. Bovine serum albumin (BSA) (Sigma-Aldrich, catalog number: V900933)
2. 6× Protein loading buffer (TransGen Biotech, catalog number: DL101-02)
3. Premixed protein marker (Takara, catalog number: 3595A)
3. Sodium chloride (NaCl) (Damao Chemical Reagent Factory, CAS number: P000967)
4. Tryptone (Oxoid, catalog number: LPOO24)
5. Yeast extract (Oxoid, catalog number: LP0021)
6. Kanamycin (Aladdin, catalog number: K103024)
7. Ampicillin (Aladdin, catalog number: A102050)
8. Isopropyl-β-D-1-thiogalactopyranoside (IPTG) (Sangon Biotech, catalog number: A100487-0025)
9. Tris(2-carboxyethyl)phosphine (TCEP) (Thermo Fisher Scientific, catalog number: T2556)
10. Sodium dihydrogen phosphate (NaH2PO4) (Sangon Biotech, catalog number: A501726)
11. Disodium hydrogen phosphate (Na2HPO4) (Sangon Biotech, catalog number: A501727)
12. Potassium dihydrogen phosphate (KH2PO4) (Sangon Biotech, catalog number: A600445)
13. Imidazole (Sigma-Aldrich, catalog number: V900153)
14. Potassium chloride (KCl) (Aladdin, catalog number: P112134)
15. Sodium sulfate (Na2SO4) (Sigma-Aldrich, catalog number: 238597)
16. Ethanolamine (Aladdin, catalog number: E103810)
17. Acetic acid (Sangon Biotech, catalog number: A501931)
18. Sodium hydroxide (NaOH) pellets (Damao Chemical Reagent Factory, catalog number: P000880)
19. Hydrochloric acid (HCl) (Cologne Chemical Reagent Factory, catalog number: LC149502)
20. Acetic acid (Sangon Biotech, catalog number: A501931)
21. Tris-base (Biofroxx, catalog number: 1115GR500)
22. Arginine (Sangon Biotech, catalog number: A600205)
23. Guanidine hydrochloride (GdnHCl) (Aladdin, catalog number: G108676)
24. Ethanol (EtOH) (Damao Chemical Reagent Factory, CAS number: 64-17-5)
25. YoungPAGETM MES SDS running buffer powder (GenScript, catalog number: M00677)
26. SOC medium (Sangon Biotech, catalog number: B540119)
Solutions
1. LB medium (see Recipes)
2. Kanamycin stock (see Recipes)
3. Ampicillin stock (see Recipes)
4. 1 M IPTG (see Recipes)
5. Binding buffer (see Recipes)
6. Elution buffer (see Recipes)
7. 100 mM PBS (see Recipes)
8. 1 M NaH2PO4 (see Recipes)
9. 1 M Na2HPO4 (see Recipes)
10. Coupling buffer (see Recipes)
11. 1 M ethanolamine (see Recipes)
12. 0.1 M acetate buffer (see Recipes)
13. 0.1 M Tris-HCl buffer (see Recipes)
14. 0.5 M arginine solution (see Recipes)
15. 6 M GdnHCl (see Recipes)
16. 0.1 M NaOH (see Recipes)
17. 20% ethanol (see Recipes)
Recipes
1. LB medium
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| NaCl | 10 g/L | 10 g |
| Tryptone | 10 g/L | 10 g |
| Yeast extract | 5 g/L | 5 g |
| Double-distilled water (ddH2O) | n/a | To 1,000 mL |
| Total | n/a | 1,000 mL |
Sterilize using an autoclave at 121 °C for 15 min.
2. Kanamycin stock
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Kanamycin | 50 mg/mL | 0.5 g |
| ddH2O | n/a | To 10 mL |
| Total | n/a | 10 mL |
Sterilize by filtering with a 0.22 μm syringe filter. Store in 1 mL aliquots at -20 °C.
3. Ampicillin stock
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Ampicillin | 100 mg/mL | 1 g |
| ddH2O | n/a | To 10 mL |
| Total | n/a | 10 mL |
Sterilize by filtering with a 0.22 μm syringe filter. Store in 1 mL aliquots at -20 °C.
4. 1 M IPTG
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| IPTG | 1 M | 2.38 g |
| ddH2O | n/a | To 10 mL |
| Total | n/a | 10 mL |
Sterilize by filtering with a 0.22 μm syringe filter. Store in 1 mL aliquots at -20 °C.
5. Binding buffer (pH 7.4)
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Na2HPO4 | 15.6 mM | 2.22 g |
| KH2PO4 | 4.4 mM | 0.6 g |
| NaCl | 500 mM | 29.22 g |
| Imidazole | 30 mM | 2.04 g |
| ddH2O | n/a | To 1,000 mL |
| Total | n/a | 1,000 mL |
Adjust pH to 7.4 with 12 M HCl. Filter through a 0.22 μm hydrophilic membrane.
6. Elution buffer (pH 7.4)
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Na2HPO4 | 15.6 mM | 2.22 g |
| KH2PO4 | 4.4 mM | 0.6 g |
| NaCl | 500 mM | 29.22 g |
| Imidazole | 500 mM | 34.04 g |
| ddH2O | n/a | To 1,000 mL |
| Total | n/a | 1,000 mL |
Adjust pH to 7.4 with 12 M HCl. Filter through a 0.22 μm hydrophilic membrane.
7. 100 mM PBS (pH 7.0)
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Na2HPO4 | 94.7 mM | 13.44 g |
| KH2PO4 | 5.3 mM | 0.72 g |
| NaCl | 137 mM | 8 g |
| KCl | 2.7 mM | 0.2 g |
| ddH2O | n/a | To 1,000 mL |
| Total | n/a | 1,000 mL |
Adjust pH to 7.0 with 12 M HCl. Filter through a 0.22 μm hydrophilic membrane.
8. 1 M NaH2PO4
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| NaH2PO4 | 1 M | 59.99 g |
| ddH2O | n/a | To 500 mL |
| Total | n/a | 500 mL |
9. 1 M Na2HPO4
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Na2HPO4 | 1 M | 70.98 g |
| ddH2O | n/a | To 500 mL |
| Total | n/a | 500 mL |
10. Coupling buffer (pH 10.0)
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Na2SO4 | 250 mM | 35.51 g |
| 1 M NaH2PO4 | 6.8 mM | 6.8 mL |
| 1 M Na2HPO4 | 93.2 mM | 93.2 mL |
| ddH2O | n/a | To 1,000 mL |
| Total | n/a | 1,000 mL |
Adjust pH to 8.0 with extra 1 M Na2HPO4. Adjust pH from 8.0 to 10.0 with 1 M NaOH. Filter through a 0.22 μm hydrophilic membrane.
11. 1 M ethanolamine (pH 8.0)
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Ethanolamine | 1 M | 12.1 mL |
| ddH2O | n/a | To 200 mL |
| Total | n/a | 200 mL |
Adjust pH to 8.0 with 12 M HCl. Filter through a 0.22 μm hydrophilic membrane.
12. 0.1 M acetate buffer (pH 4.0)
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Acetic acid | 0.1 M | 571.9 μL |
| NaCl | 0.5 M | 2.92 g |
| ddH2O | n/a | To 100 mL |
| Total | n/a | 100 mL |
Adjust pH to 4.0 with 1 M NaOH. Filter through a 0.22 μm hydrophilic membrane.
13. 0.1 M Tris-HCl buffer (pH 8.0)
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Tris-Base | 0.1 M | 1.21 g |
| NaCl | 0.5 M | 2.92 g |
| ddH2O | n/a | To 100 mL |
| Total | n/a | 100 mL |
Adjust pH to 8.0 with 12 M HCl. Filter through a 0.22 µm hydrophilic membrane.
14. 0.5 M arginine solution (pH 3.8)
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Arginine | 0.5 M | 17.42 g |
| ddH2O | n/a | To 200 mL |
| Total | n/a | 200 mL |
Adjust pH to 3.8 with 12 M HCl. Filter through a 0.22 μm hydrophilic membrane.
15. 6 M GdnHCl (pH 2.0)
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Guanidine hydrochloride | 6 M | 289.5 g |
| ddH2O | n/a | To 500 mL |
| Total | n/a | 500 mL |
Adjust pH to 2.0 with 12 M HCl. Filter through a 0.22 μm hydrophilic membrane.
16. 0.1 M NaOH
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| NaOH | 0.1 M | 2 g |
| H2O | n/a | To 500 mL |
| Total | n/a | 500 mL |
Filter through a 0.22 μm hydrophilic membrane.
17. 20% ethanol
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Ethanol | 20% (v/v) | 200 mL |
| ddH2O | n/a | To 1,000 mL |
| Total | n/a | 1,000 mL |
Filter through a 0.22 μm hydrophilic membrane.
Laboratory supplies
1. HisTrapTM HP column (Cytiva, catalog number: 17524802)
2. Epoxy-activated Seplife® FF resin (Sunresin, catalog number: A40309010)
3. Spectrophotometer cuvettes (Sangon, catalog number: F511002-0001)
4. Empty spin columns (Biocomma, catalog number: 007400)
5. 1 mL empty chromatography column (NanoMicro, catalog number: 10000077022X)
6. 0.2 mL PCR tubes (Axygen, catalog number: PCR-2CP-RT-C)
7. 1.5 mL microcentrifuge tubes (Axygen, catalog number: MCT-150-C)
8. 2 mL microcentrifuge tubes (Axygen, catalog number: MCT-200-C)
9. 15 mL centrifuge tubes (Corning, catalog number: 430791)
10. 50 mL centrifuge tubes (Corning, catalog number: 430829)
11 0.22 μm syringe filters (Pall Corporation, catalog number: 4652)
12. 20 mL syringes (double-dove, catalog number: HX-SLZSQ-SG-20ML/16)
13. 0.22 μm hydrophilic membrane filter (Millipore Sigma, catalog number: GSWP04700)
14. Amicon Ultra centrifugal filter units (3 K MWCO) (Millipore Sigma, catalog number: UFC900396)
15. Amicon Ultra centrifugal filter units (10 K MWCO) (Millipore Sigma, catalog number: UFC901096)
16. YoungPAGETM Precast gel (GenScript, catalog number: M00928)
17. SnakeSkinTM dialysis tubing, 3.5 K MWCO (Thermo Fisher Scientific, catalog number: 88244)
Equipment
1. Pipetting system (Gilson, model: PIPETMAN Classic Starter Kit)
2. Shaking incubator (New Brunswick Scientific, model: Innova44)
3. Biochemical incubator (Thermo Fisher Scientific, model: IMH100-S)
4. Spectrophotometer (Youke Instrument, model: UV755B)
5. Electronic analytical balance (Mettler Toledo, model: MS204TS)
6. Autoclave (Hirayama, model: HVE-50)
7. -80 °C freezer (Thermo Fisher Scientific, model: 995)
8. Clean bench (Donglian, model: DL-CJ-2NDII)
9. Ultrasonic homogenizer (Scientz, model: JY92-)
10. Centrifuge for 15 and 50 mL tubes (Eppendorf, model: 5810R)
11. Centrifuge for 1.5 and 2 mL tubes (Eppendorf, model: 5425R)
12. Electrophoresis equipment (Bio-Rad, model: Mini-PROTEAN® Tetra Cell Systems)
13. pH meter (Mettler Toledo, model: FE28-standard)
14. Mini rotator (Gilson, model: Roto-MiNi Plus)
15. AKTA Purifier chromatography system (GE Healthcare, model: AKTA Purifier 10)
16. Protein staining instrument (GenScript, model: eStain L1)
17. Vacuum filtration system (Ilmvac GmbH, model: 120788)
18. Dry bath incubator (Coyote, model: H2O3-100C)
Software and datasets
1. SnapGene® (SnapGene, https://www.snapgene.com/)
2. ImageJ (National Institutes of Health DNA, https://imagej.net/ij/)
3. Unicorn for AKTA purifier (GE Healthcare, https://www.cytivalifesciences.com/en/us/shop/chromatography/software/unicorn-7-p-05649)
Procedure
A. Protein expression
1. Transform E. coli BL21(DE3) bacterial strain with the pET30a-6 × His-SpyFixer (kanamycin-resistant) plasmid and pET32a-Z domain-SpyTag002-6×His (ampicillin-resistant) plasmid for protein production. The plasmid maps and corresponding protein sequences are shown in Figure 1. Use the standard heat-shock method for transformation:
a. Add 1–50 ng of plasmid DNA to 100 μL of chemically competent cells. Mix by gentle tapping and place on ice.
b. Incubate the cells on ice for 30 min.
c. Transfer the cells to a 42 °C biochemical incubator for exactly 90 s.
d. Transfer the cells to ice for 2 min.
e. Add 900 μL of SOC medium to the cells.
f. Incubate the cells on a shaking incubator (220 rpm) at 37 °C for 45 min.
g. Pipette 200 µL of transformed cell suspension onto an LB agar plate with kanamycin and spread it using a sterile spreader.
h. Incubate the plate overnight at 37 °C.
i. Perform colony PCR and Sanger sequencing to select an E. coli colony transformed with a plasmid vector containing the insert fragment.
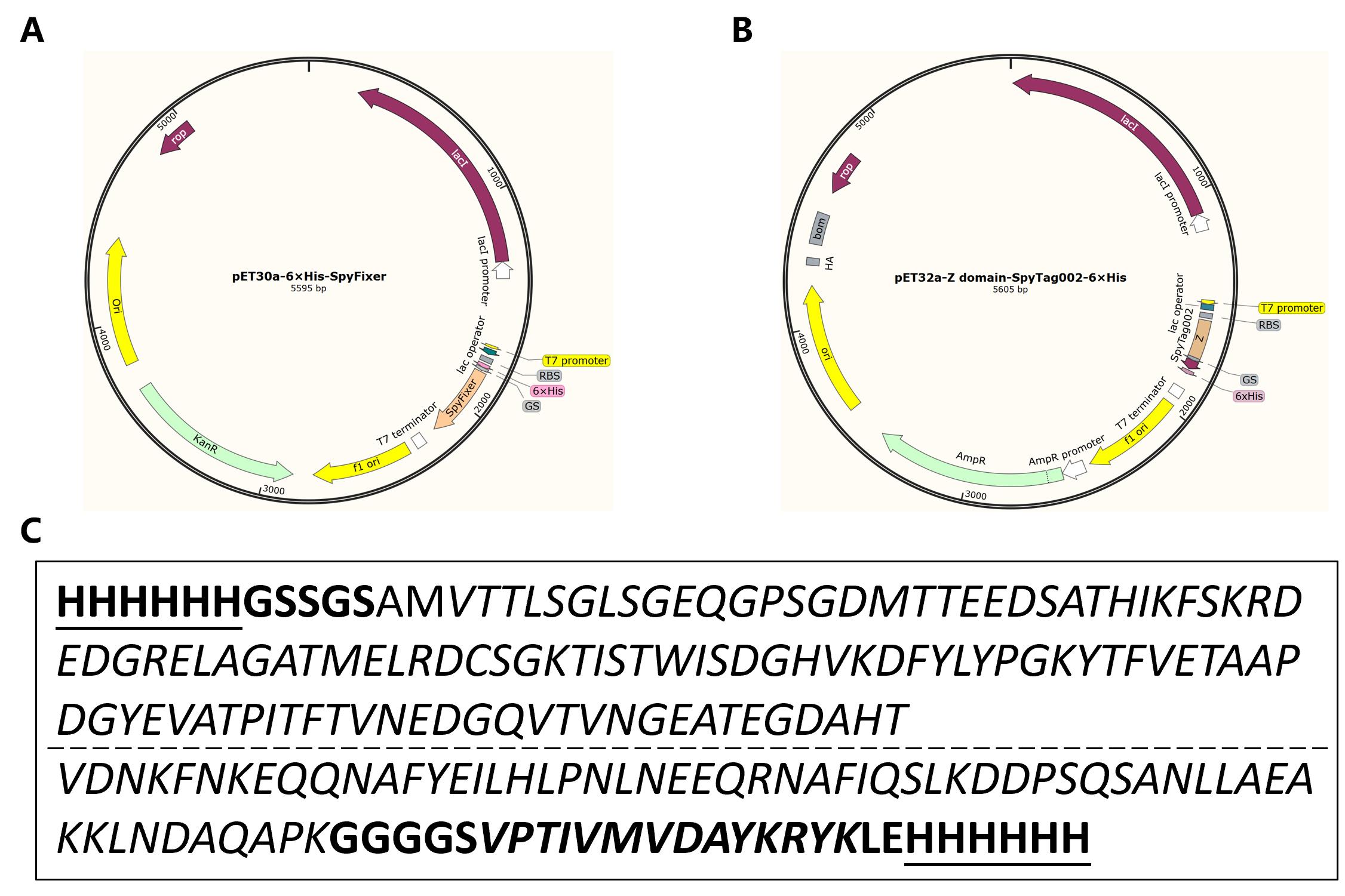
Figure 1. Plasmid maps and protein sequences. (A) Plasmid map of pET30a-6×His-SpyFixer. (B) Plasmid map of pET32a-Z domain-SpyTag002-6×His. (C) Sequences of fusion proteins. The sequence of the 6-His tag is underlined, while the sequence of SpyTag002 is in bold and italics. The SpyFixer sequence is in italics and not bold (top), and the Z domain sequence is in italics and not bold (bottom).
2. Inoculate a single bacterial colony into 10 mL of LB medium supplemented with the appropriate antibiotics (50 μg/mL kanamycin for pET30a-6×His-SpyFixer plasmid and 100 μg/mL ampicillin for pET32a-Z domain-SpyTag002-6×His) in a 50 mL centrifuge tube. Grow overnight at 37 °C with shaking at 220 rpm.
3. The next day, transfer the 10 mL primary culture to 500 mL of LB medium containing appropriate antibiotics in a 2 L conical flask. Grow at 37 °C with shaking at 220 rpm until OD600 reaches 0.4–0.6.
Note: Typically, it takes 1.5–2 h to reach an OD600 of 0.4.
4. Once in the OD range, add IPTG to a final concentration of 0.2 mM and let bacteria grow at 18 °C for 24 h.
Note: For SpyFixer expression, an optional step includes inducing with 0.42 mM IPTG and growing for 4 h at 30 °C at 220 rpm.
5. After induction, pipette 1 mL of the culture to measure the final OD600. Then, transfer the remaining culture into 50 mL centrifuge tubes.
6. Pellet bacteria by centrifugation at 4,000× g for 20 min at 4 °C. Freeze the bacterial cell pellet at -80 °C until use.
B. Protein preparation
1. Thaw one tube of bacterial cell pellet on ice (6×His-SpyFixer or Z domain-SpyTag002-6×His).
2. Resuspend the pellet thoroughly in ice-cold binding buffer to a final OD600 of 50 by pipetting up and down using a 1 mL micropipette tip.
Note: For example, if the cells are harvested at an OD600 of 5, resuspend the cell pellet from each 10 mL of culture in 1 mL of binding buffer.
3. Bacterial cell lysis: Disrupt the suspension solution on ice with an ultrasonic homogenizer to allow for complete lysis (settings: 3 s ON, 3 s OFF, power = 195 watts, amplitude probe = 6 mm). Complete lysis will be evident as the lysate becomes light brown in color.
Note: It is recommended to place the centrifuge tube in a beaker filled with an ice-water mixture to keep the samples cold during sonication.
4. Centrifuge the lysate at 15,000× g for 20 min at 4 °C to pellet insoluble material and debris. Carefully transfer the supernatant to a new pre-chilled centrifuge tube on ice.
5. Filter the supernatant of cell lysate through a 0.22 μm syringe filter to remove particulate matter.
6. Perform Ni-NTA affinity purification using a fast protein liquid chromatographic system (AKTA purifier) with a 5 mL HisTrapTM HP column connected in series.
a. Pre-equilibrate the column with 10 column volumes (CVs) of binding buffer.
b. Load the filtered supernatant onto the pre-equilibrated column at a flow rate of 1 mL/min to ensure efficient binding of His-tagged proteins.
7. Wash the column with binding buffer until the absorbance at 280 nm stabilizes to remove nonspecifically bound proteins.
8. Elute the target protein with 0%–100% elution buffer for 30 CVs while monitoring absorbance at 280 nm to identify elution peaks. Collect elution fractions in 50 mL centrifuge tubes and store at 4 °C.
9. Clean the column after elution by washing with elution buffer to remove any residual protein bound to the column. Wash with 5 CVs of ddH2O and store the column in 20% ethanol.
10. Prepare protein samples for SDS-PAGE analysis.
a. Mix an aliquot of each elution fraction with 6× protein loading buffer at a 5:1 volume ratio in 0.2 mL PCR tubes (e.g., mix 20 μL of elution fraction with 4 μL of 6× protein loading buffer).
b. Heat the mixture at 98 °C for 10 min in a dry bath incubator (or water bath) to denature proteins.
Note: Ensure the tubes are tightly sealed to prevent evaporation.
c. Place 4%–20% YoungPAGETM Precast gel in YoungPAGETM MES SDS running buffer and load 8 μL of protein samples on the gel.
d. Run the gel at 180 V for 30 min or until the tracking dye reaches the bottom of the gel.
e. Visualize the SDS-PAGE results using the eStain L1 protein staining system, following the manufacturer’s instructions.
11. Pool the fractions containing the purified target protein (as determined by SDS-PAGE analysis) and concentrate using Amicon® ultra centrifugal filters (3 kDa or 10 kDa cutoff) at 3,220× g in an Eppendorf 5810 R centrifuge at 4 °C until the final volume is ~1–2 mL.
12. Dialyze the concentrated protein overnight at 4 °C using dialysis tubing (3.5 kDa MWCO) with 2 L of ice-cold coupling buffer for SpyFixer or 2 L of ice-cold 100 mM PBS for Z domain-SpyTag002 in the beaker with gentle stirring.
Note: To ensure successful immobilization onto the resin in this protocol, the chosen solution should align with the recommended conditions.
C. Preparation of SpyFixer-modified resin
Note: This procedure uses 250 μL of resin as an example. Adjust the volumes of all solutions proportionally for larger or smaller resin volumes. All buffer washes are performed by centrifugation at 135× g for 1 min at 4 °C.
1. Add 250 μL of ddH2O to empty spin columns. Mark the desired resin volume (250 μL) on the column tube using a marker. Discard the water by centrifugation at 135× g for 30 s at 4 °C.
2. Transfer epoxy-activated Seplife® FF resin into the prepared spin columns to allow the resin to settle naturally until the volume reaches the marked line (250 μL). Wash the resin five times with 500 μL of ddH2O followed by five washes with coupling buffer.
3. Add a final concentration of 1 mM TCEP to the dialyzed SpyFixer at 25 °C for 30 min to reduce the SpyFixer.
Add 25 mg of reduced SpyFixer to the 250 μL of prepared epoxy-activated Seplife® FF. Shake the resin slurry at 14 rpm for 12 h at 25 °C.
4. After standing for 5 min, collect the resin by centrifugation at 135× g for 1 min at 4 °C and wash it five times with 2 CVs of coupling buffer.
5. Block unreacted epoxy groups by incubating the resin in 2 CVs of 1 M ethanolamine solution at 100 rpm for 12 h at 37 °C.
6. After 12 h, wash the resin three times each with 2 CVs of 0.1 M acetate buffer and 2 CVs of 0.1 M Tris-HCl buffer in alternating cycles.
7. Store SpyFixer-modified resin in 2 CVs of 20% ethanol at 4 °C.
D. Preparation of SpyFixer/Z domain–modified resin
Note: All buffer washes are performed by centrifugation at 135× g for 1 min at 4 °C.
1. Wash 250 μL of SpyFixer-modified resin five times with 500 μL of 100 mM PBS.
2. Incubate the SpyFixer-modified resin (250 μL) with 15 mg of dialyzed Z domain-SpyTag002 in 400 μL of coupling buffer at 14 rpm for 2 h at 25 °C. If the volume of protein solution is less than 400 μL, adjust it to the final volume using coupling buffer.
3. After 12 h, collect the resin by centrifugation at 135× g for 1 min at 4 °C.
4. Wash the resin five times with 100 mM PBS at room temperature to remove any unbound Z domain-SpyTag002.
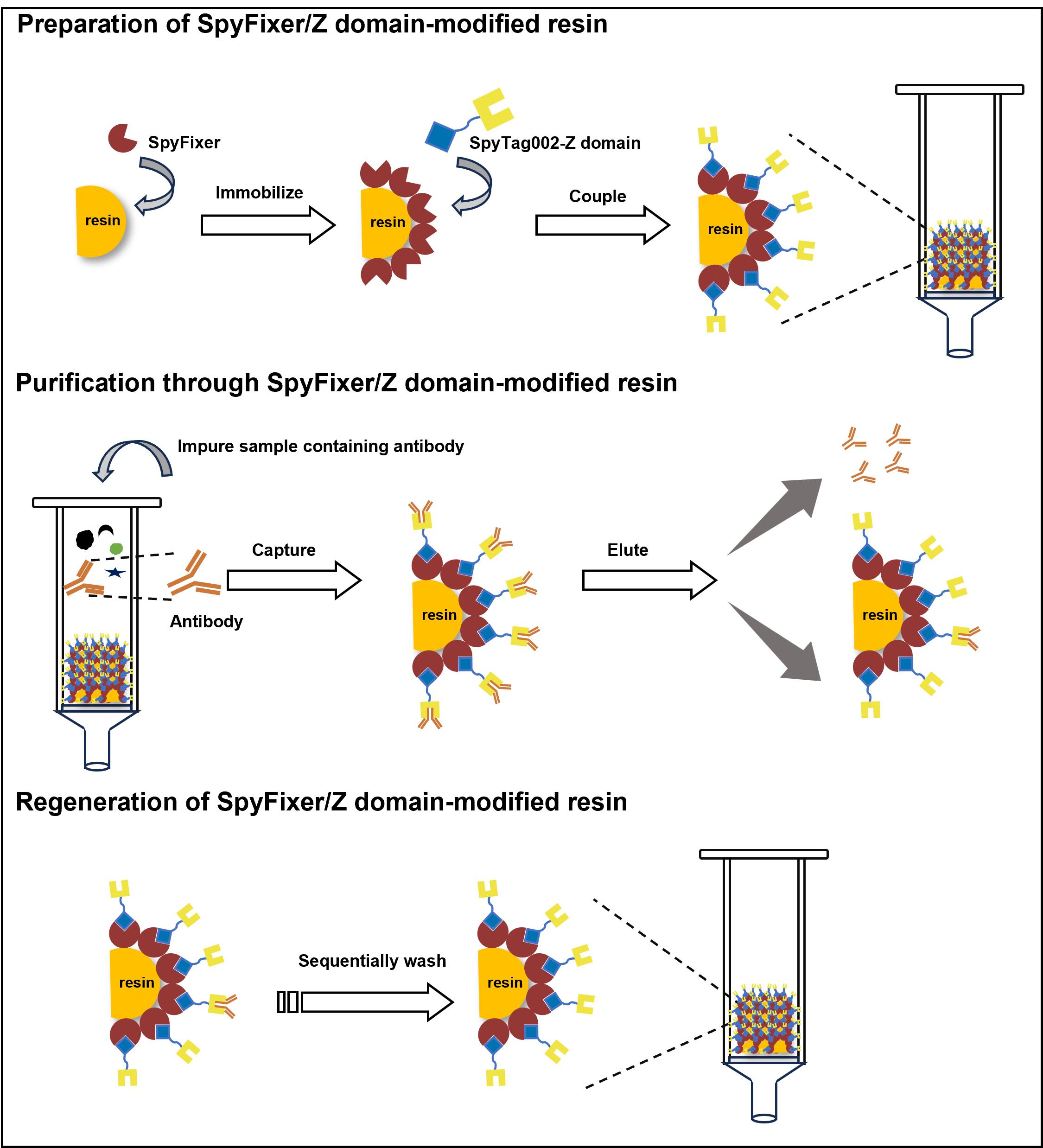
5. Store SpyFixer/Z domain–modified resin (Figure 2) in 2 CVs of 20% ethanol at 4 °C until use.
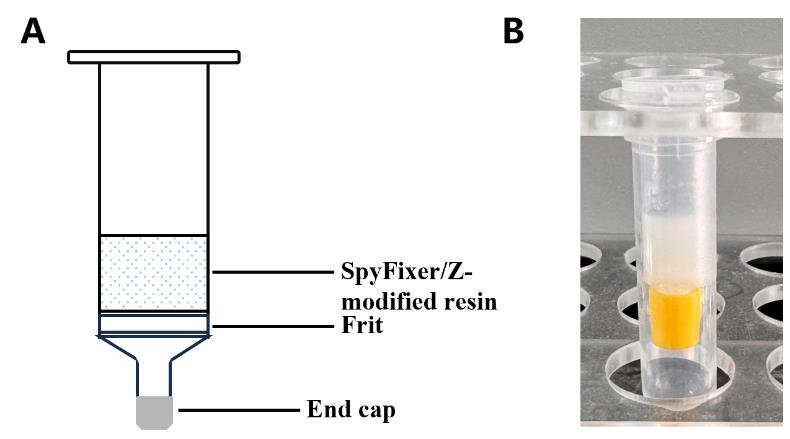
Figure 2. Diagram of a SpyFixer/Z domain–modified resin affinity column assembly. (A) Schematic and (B) photo depicting column apparatus.
E. Purification through SpyFixer/Z domain–modified resin
Note: All buffer washes are performed by centrifugation at 135× g for 1 min at 4 °C.
1. Incubate SpyFixer/Z domain–modified resin with antibody samples intended for purification and shake the mixture at 14 rpm for 2 h at 25 °C.
2. After 2 h, separate the resin-bound hIgG from the supernatant by centrifuging at 135× g for 1 min at 4 °C.
3. Wash the resin five times with 500 μL of 100 mM PBS to remove nonspecifically bound proteins.
4. Elute the target antibody by passing 1.5 CVs of 0.5 M arginine solution through the resin, repeating this step eight times or more as needed. Collect each elution fraction by centrifugation at 135× g for 1 min at 4 °C and transfer each elution fraction to new microcentrifuge tubes (e.g., 1.5 mL tube)
5. Analyze the collected target protein fractions on a 4%–20% SDS-PAGE gel as described in section C (Figure 3).
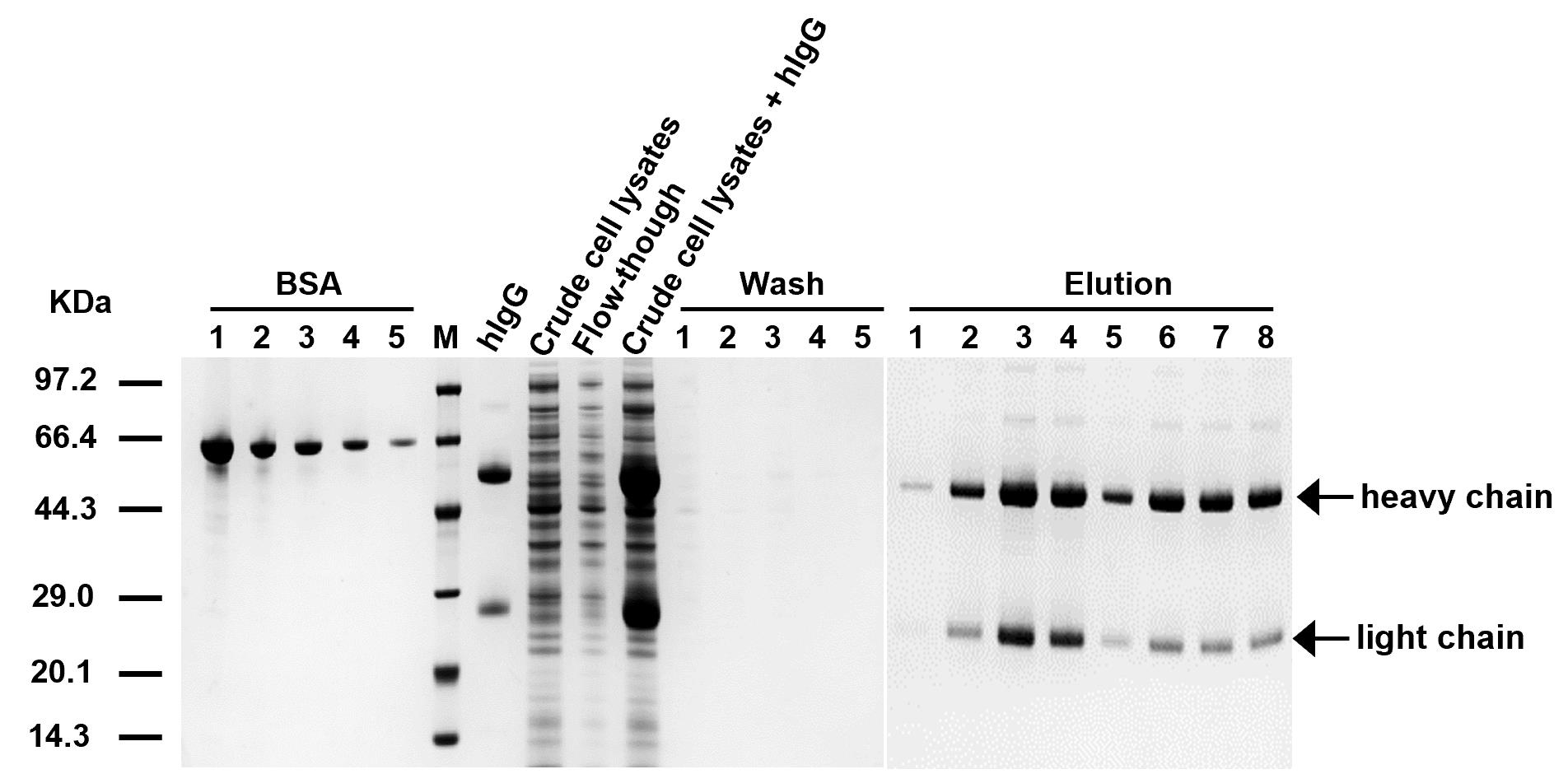
Figure 3. SDS-PAGE analysis of purification steps using SpyFixer/Z domain–modified resin. (1) Human immunoglobulin G (hIgG) (purified hIgG), (2) crude cell lysates (E. coli lysates), (3) flowthrough (unbound proteins), (4) crude cell lysates + hIgG (hIgG mixed with E. coli lysates) (5) wash (removal of nonspecific binding), and (6) elution (purified target antibody). BSA standard concentrations 1–5 were 0.5, 0.25, 0.125, 0.0625, and 0.03125 mg/mL, respectively. Figure from Xiang et al. [1]
F. Regeneration of SpyFixer/Z domain–modified resin (Figure 4)
Note: All buffer washes are performed by centrifugation at 135× g for 1 min at 4 °C.
1. After elution, wash the resin three times with 500 μL of 6 M GdnHCl at room temperature to remove residual bound proteins or contaminants.
2. Follow with three washes using 500 μL of 100 mM PBS, pH 7.4 to neutralize and rinse away any residual GdnHCl.
3. Wash the resin three times with 500 μL of 0.1 M NaOH at room temperature to effectively remove any remaining protein or contaminants.
4. Rinse the resin thoroughly with 500 μL of 100 mM PBS at room temperature to neutralize the NaOH and stabilize the resin.
5. Wash the resin extensively ten times with 500 μL of 100 mM PBS at room temperature to ensure complete removal of residual reagents.
6. Store the resin in 20% ethanol at 4 °C to maintain sterility and prolong usability.
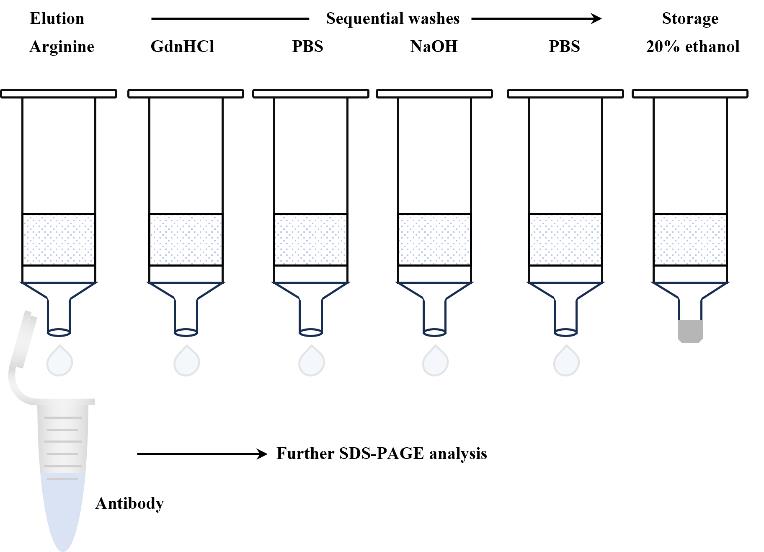
Figure 4. Scheme for regeneration steps of the SpyFixer/Z domain–modified resin
Data analysis
To assess the purity and amount of the proteins, run the purified samples on an SDS-PAGE gel. The composition and protein amounts are determined densitometrically by using the software ImageJ using BSA as standard (Figure 5). A series of BSA standards at known concentrations (0.5, 0.25, 0.125, 0.0625, and 0.03125 mg/mL) are prepared and loaded alongside the protein samples. After electrophoresis, the gel is stained using the eStain L1 protein staining system. Band intensities are measured with ImageJ, and a BSA standard curve is generated by plotting known protein concentration (in x-axis) vs. band intensity (in y-axis). Protein concentration for the target protein is estimated by plotting the band intensity of the target protein (in the y-axis), determining the intersection point with the BSA standard curve, and then finding the concentration associated with that particular point (in the x-axis).
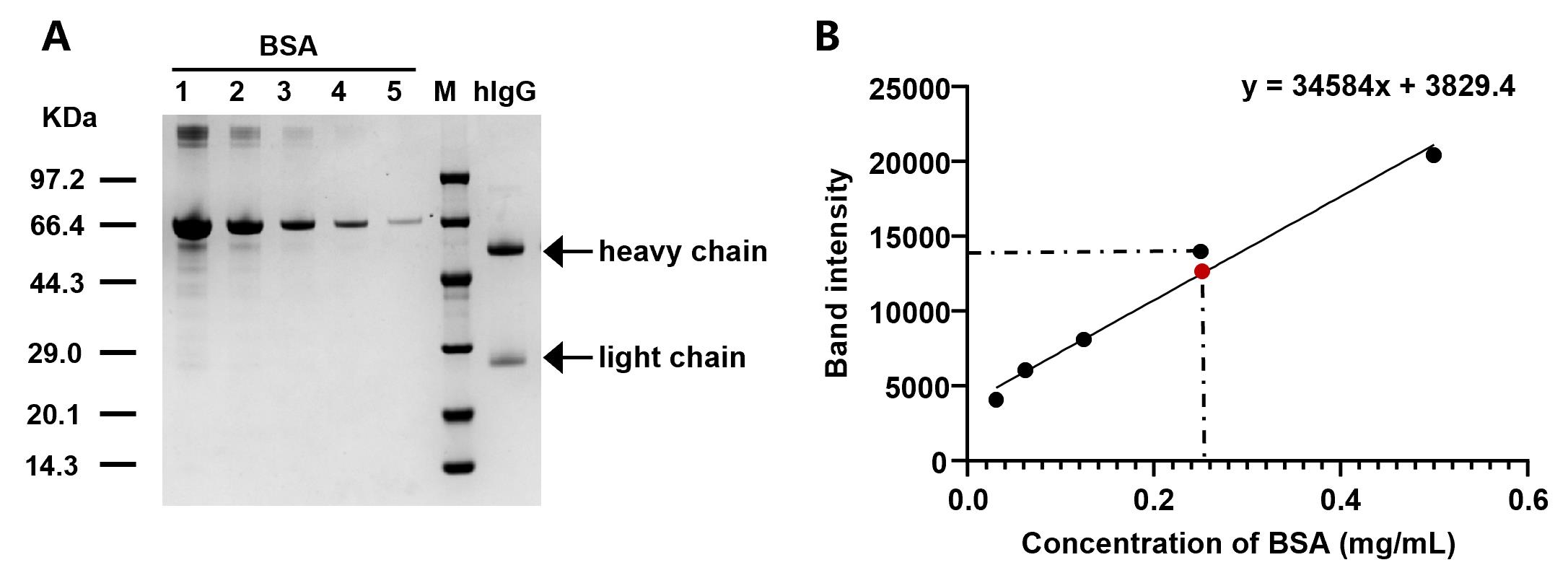
Figure 5. SDS-PAGE analysis and BSA standard curve for human immunoglobulin G (hIgG) quantification. (A) SDS-PAGE analysis of purified hIgG. BSA standard concentrations 1–5 correspond to 0.5, 0.25, 0.125, 0.0625, and 0.03125 mg/mL, respectively. (B) BSA standard curve. The black dots represent BSA band intensities at known concentrations (0.03125, 0.0625, 0.125, 0.25, and 0.5 mg/mL), which are used to generate the standard curve. The red dot represents the band intensity of the target protein (hIgG). This BSA curve and equation will be used to calculate hIgG concentration.
The SpyFixer/Z domain–modified resin enabled the purification of hIgG from cell lysate, human serum, and industrial fermentation broth with high purity (>95%). Quantitative data on antibody yield and purity are provided in Table 1, while additional details can be found in the original research paper [1].
Table 1. Purification results obtained with purified hIgG, crude cell lysates containing hIgG, human serum containing hIgG, and fermentation broth containing recombinant hIgG4. Table from Xiang et al. [1].
| Sample | hIgG loaded (mg/mL resin) | Capture efficiency (%) | Elution efficiency (%) | Purity (%) |
| Purified hIgG | 240 | 94.0 ± 2.3 | 79.5 ± 2.3 | 95.9 ± 0.5 |
hIgG mixed with E. coli lysates | 50 | >99.0 | 79.7 ± 4.7 | 95.8 ± 0.5 |
| hIgG from human serum | 17 | 96.5 ± 0.4 | 86.7 ± 9.1 | 95.3 ± 1.5 |
| Recombinant hIgG4 from fermentation broth | 14 | 94.9 ± 1.3 | 84.0 ± 2.5 | 96.0 ± 0.7 |
Purification of hIgG using the regenerated resin yielded results comparable to those obtained with new resin (Figure 6). Details can be found in the original research paper [1].
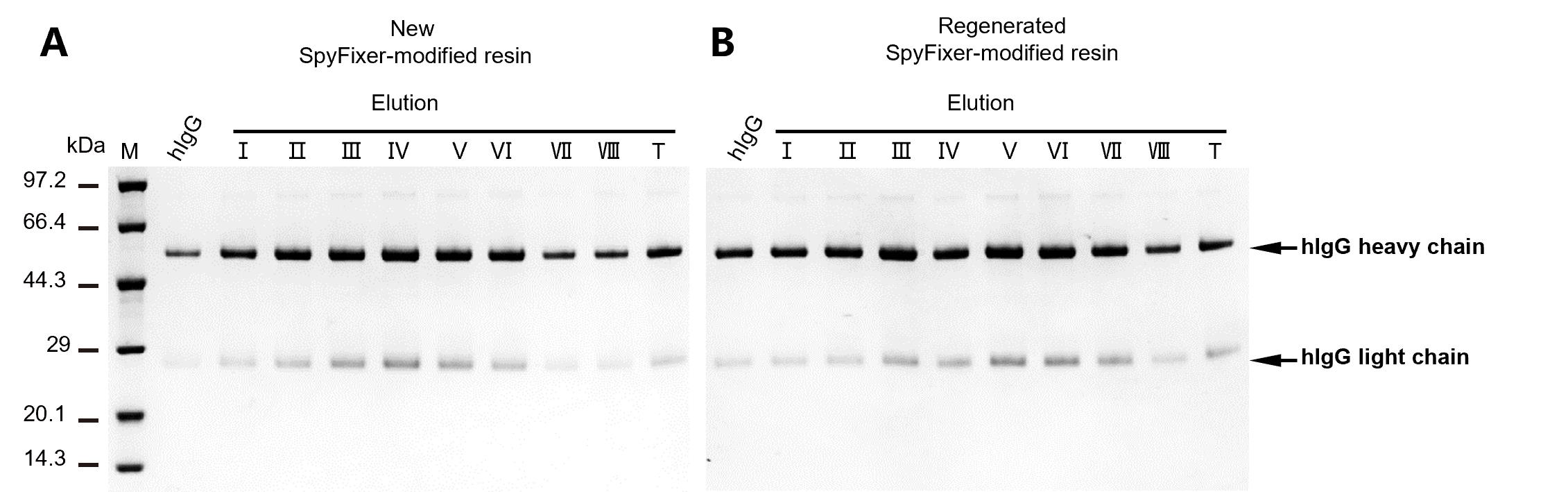
Figure 6. Reusability of SpyFixer/Z domain–modified resin. SpyFixer/Z domain–based antibody purification of human immunoglobulin G (hIgG) using new unused resin or regenerated resin. Elution fractions – and the total pooled elution (T) fraction are shown. hIgG: input of hIgG. Figure from Xiang et al. [1].
The purification process using SpyFixer/Z domain–modified resin is straightforward and cost-effective, achieving hIgG purification and reusability with comparable efficiency to commercial Protein A resin. Notably, the loading capacity of the Z domain–modified resin for hIgG exceeds 200 mg/mL of resin, which is five times higher than that of the commercial Protein A resin. Furthermore, SpyFixer and the SpyTagged Z domain can be expressed in bacteria with high yields (>100 mg/L at the shaker level), and the resin used in this protocol is cost-effective, making it a promising alternative for large-scale commercial applications. Details can be found in the original research paper [1].
Validation of protocol
This protocol or parts of it has been used and validated in the following research article:
Xiang et al [1]. SpyFixer enables efficient site-specific immobilization for protein-protein interaction analysis and antibody purification. Int J Biol Macromol (Figure 2).
General notes and troubleshooting
General notes
1. This protocol describes antibody capture and elution through centrifugation. Alternatively, a pre-packed SpyFixer/Z domain–modified resin affinity column (Figure 7) can be prepared for connection to a liquid chromatography system such as AKTA.
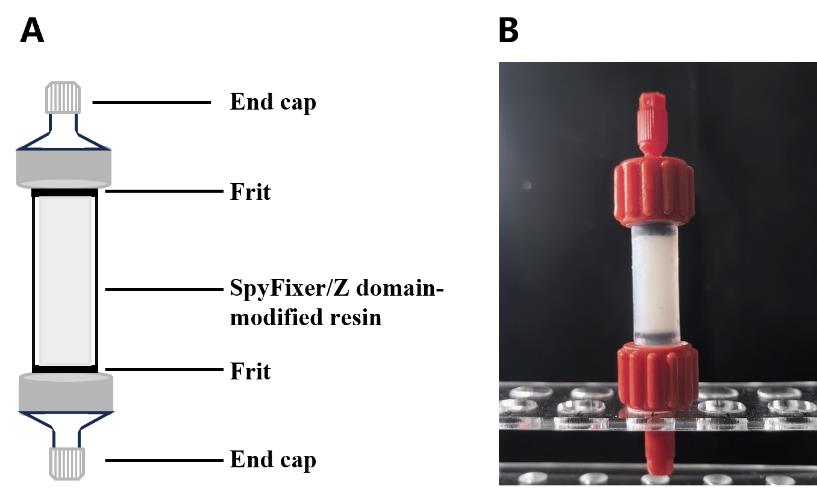
Figure 7. Diagram of a pre-packed SpyFixer/Z domain–modified resin affinity column assembly. (A) Schematic and (B) photo depicting column apparatus.
2. Keep elution fractions and protein samples on ice when appropriate to not lose activity.
3. Filter-sterilize all buffers before use and store them at 4 °C for optimal stability. Storage at room temperature is also acceptable; however, if aggregation or precipitation is observed, the buffer should be freshly prepared to ensure consistency and prevent interference in experimental results.
4. For larger quantities of harvested cells, a high-pressure homogenizer can be used as an alternative to a sonicator for cell lysis.
5. To ensure optimal binding efficiency and maintain protein activity, use freshly prepared proteins for the preparation of the antibody purification resins as described in this protocol.
6. The SpyFixer/Z domain–modified resin primarily binds to the Fc region of human IgG subclasses (IgG1, IgG2, and IgG4) with high affinity.
7. For 250 μL of resin in empty spin columns, load 250–400 μL of antibody sample. We recommend using 100 mM PBS for the antibody incubation described in this protocol, as it does not affect the Spy reaction. Additionally, this protocol enables efficient hIgG purification from E. coli cell lysate, human serum, or industrial fermentation broth.
Troubleshooting
Problem 1: Low protein yields due to cell lysis (step B3).
Possible cause: Incomplete cell lysis.
Solution: Sonicate the cell suspension for 30 min. Ensure that the suspension becomes less viscous and slightly darker.
Problem 2: Low protein immobilization efficiency (Sections C and D).
Possible cause: Loss of protein activity or partial inactivation of epoxy groups on the resin due to prolonged storage.
Solution: Use freshly prepared protein and epoxy-activated resin from a newly synthesized batch to maximize immobilization efficiency.
Problem 3: Low recovery of the target antibody (step E4).
Possible cause: Incomplete antibody release from the resin.
Solution: Increase the number of elution steps to improve antibody recovery. If recovery remains low, optimize elution conditions by modifying buffer composition, pH, and ionic strength to enhance antibody release efficiency.
Acknowledgments
Conceptualization, X.Y. (Xiaofeng Yang) and Z.L. (Zhanglin Lin); Investigation, Y.X. (Ya Xiang) and Z.L. (Zisha Lao); Writing—Original Draft, Y.X.; Writing—Review & Editing, X.Y. and Z.L.; Funding acquisition, X.Y; Supervision, X.Y. and Z.L.
This study was supported by the National Key Research and Development Program of China (2022YFC2104800). The protocol described here was adapted from previously reported studies by the authors [1].
Competing interests
X.Y., Z.L., Y.X. and Z.L. have patents pending to China National Intellectual Property Administration.
References
- Xiang, Y., Lao, Z., Lin, Z., Yang, X. (2024). SpyFixer enables efficient site-specific immobilization for protein-protein interaction analysis and antibody purification. Int J Biol Macromol. 138548. https://doi.org/10.1016/j.ijbiomac.2024.138548.
- Monoclonal Antibody Therapy Market Size, Share & Industry Analysis, By Type (Human mAb, Humanized mAb, Chimeric mAb, and Murine mAb), By Application (Cancer, Autoimmune Diseases, and Others), By Distribution Channel (Hospital Pharmacy, Retail Pharmacy, and Online Pharmacy), and Regional Forecast, 2024–2032. (Accessed on 1/21/2025, https://www.fortunebusinessinsights.com/monoclonal-antibody-therapy-market-102734).
- Luo, S., Zhang, B. (2024). Benchmark glycan profile of therapeutic monoclonal antibodies produced by mammalian cell expression systems. Pharm Res. 41(1): 29–37. https://doi.org/10.1007/s11095-023-03628-4.
- Gagnon, P. (2012). Technology trends in antibody purification. J Chromatogr A. 1221: 57–70. https://doi.org/10.1016/j.chroma.2011.10.034.
- Ko, K.Y., Ahn, D.U. (2007). Preparation of immunoglobulin Y from egg yolk using ammonium sulfate precipitation and ion exchange chromatography. Poult Sci. 86(2): 400–407. https://doi.org/10.1093/ps/86.2.400.
- D. Low, D., O’Leary, R., Pujar, N.S. (2007). Future of antibody purification. J Chromatogr B. 848(1): 48–63. https://doi.org/10.1016/j.jchromb.2006.10.033.
- Rodrigo, G., Gruvegård, M., Van Alstine, J.M. (2015). Antibody fragments and their purification by protein L affinity chromatography. Antibodies. 4(3): 259–277. https://doi.org/10.3390/antib4030259.
- Zarrineh, M., Mashhadi, I.S., Farhadpour, M., Ghassempour, A. (2020). Mechanism of antibodies purification by protein A. Anal Biochem. 609: 113909. https://doi.org/10.1016/j.ab.2020.113909.
- Ramos-de-la-Peña, A.M., González-Valdez, J., Aguilar, O. (2019). Protein A chromatography: Challenges and progress in the purification of monoclonal antibodies. J Sep Sci. 42(9): 1816–1827. https://doi.org/10.1002/jssc.201800963.
- Sugiura, T., Imagawa, H., Kondo, T. (2000). Purification of horse immunoglobulin isotypes based on differential elution properties of isotypes from protein A and protein G columns. J Chromatogr. B 742(2): 327–334. https://doi.org/10.1016/S0378-4347(00)00177-8.
- Wang, W., Hao, D., Ge, J., Zhao, L., Huang, Y., Zhu, K., Wu, X., Su, Z., Yu, R., Ma, G. (2019). A minimalist peptide ligand for IgG by minimizing the binding domain of protein A. Biochem Eng J. 151: 107327. https://doi.org/10.1016/j.bej.2019.107327.
- Löfblom, J., Feldwisch, J., Tolmachev, V., Carlsson, J., Ståhl, S., Frejd, F.Y. (2010). Affibody molecules: Engineered proteins for therapeutic, diagnostic and biotechnological applications. FEBS Lett. 584(12): 2670–2680. https://doi.org/10.1016/j.febslet.2010.04.014.
- Zakeri, B., Fierer, J.O., Celik, E., Chittock, E.C., Schwarz-Linek, U., Moy, V.T., Howarth, M. (2012). Peptide tag forming a rapid covalent bond to a protein, through engineering a bacterial adhesin. Proc Natl Acad Sci USA. 109(12): E690–E697. https://doi.org/10.1073/pnas.1115485109.
Article Information
Publication history
Received: Jan 21, 2025
Accepted: Mar 19, 2025
Available online: Apr 3, 2025
Published: Apr 20, 2025
Copyright
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Yang, X., Lin, Z., Xiang, Y. and Lao, Z. (2025). Antibody Purification Using Spy Chemistry. Bio-protocol 15(8): e5286. DOI: 10.21769/BioProtoc.5286.
Category
Biochemistry > Protein > Isolation and purification
Biochemistry > Protein > Interaction > Protein-protein interaction
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









