- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
A Robust and Easy Protein Purification Method Using SpyDock-Modified Resin
Published: Vol 15, Iss 8, Apr 20, 2025 DOI: 10.21769/BioProtoc.5270 Views: 2007
Reviewed by: Alessandro DidonnaNuttavut KosemSneha Ray

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
Expression and Purification of the Human Voltage-Gated Proton Channel (hHv1)
Emerson M. Carmona [...] Luis G. Cuello
Jun 20, 2025 1953 Views
Thermus thermophilus CRISPR Cas6 Heterologous Expression and Purification
Junwei Wei [...] Yingjun Li
Jul 20, 2025 2137 Views
Prokaryotic Expression and Purification of the hSox2-HMG Domain
Lijie Yang [...] Jingjun Hong
Aug 20, 2025 2324 Views
Abstract
Protein purification is a critical step in both life sciences and biomanufacturing. Traditional affinity chromatography (AC) methods, including His-tag-based purification, provide high-purity proteins but are limited by the high cost of resins and the need for additional tag-removal steps. In this protocol, we present a reusable SpyDock-modified epoxy resin coupled with a pH-inducible self-cleaving intein for direct purification of proteins with authentic N-termini. This method enables efficient protein purification from cell lysates, achieving high purity (>90%) and yields comparable to the His-tag approach, without requiring tag removal. The SpyDock-modified resin protocol is robust, easy to implement, and cost-effective, making it suitable for both research and large-scale industrial applications.
Key features
• This protocol offers a robust and straightforward method for purifying proteins with authentic N-termini, eliminating the need for additional tag removal steps.
• The approach achieves higher purity and comparable yields to the commercial His-tag method.
• The SpyDock-modified epoxy resin is easy to prepare, cost-effective, and reusable.
Keywords: Protein purificationGraphical overview
Background
Protein purification constitutes a foundational process in life sciences and biomanufacturing. It plays a pivotal role in producing high-quality proteins used in research, diagnostics, and therapeutic applications. Among the diverse array of available methods, affinity chromatography remains a dominant technology due to its high specificity and efficiency, which rely on specific interactions between target proteins and immobilized ligands [1]. However, the use of affinity tags, such as the His-tag, can present challenges, including potential disruptions to protein solubility, structure, and function, as well as complications from metal ion leakage and immunogenicity [2,3]. Moreover, the subsequent removal of tags via proteolysis can reduce yields and introduce additional costs [4]. These limitations have, in turn, prompted the development of more efficient, robust, and cost-effective purification strategies.
The SpyTag‐SpyCatcher protein ligase system has recently emerged as a new tool for protein purification, owing to its unique capacity to form stable and highly specific covalent complexes [5]. While previous methods utilizing the SpyTag-SpyCatcher system have been described, they still face limitations, such as the need for chemical peptide synthesis and challenges related to the non-reusability of the covalent linkage [6]. More recently, the engineered SpyDock (13.6 kDa) has provided a non-reactive version of SpyCatcher immobilized on SulfoLink resin, which offers advantages. However, its application has been constrained by the need for an additional SpyTag in the protein of interest (POI), oxidation-sensitive thioether bonds, and high resin costs, limiting its scalability for industrial use [3].
Here, we present a robust and easy protein purification protocol utilizing SpyDock-modified resin to directly purify SpyTag-fusion proteins from crude cell lysates. This method achieves high purities (>90%) and comparable yields to the conventional His-tag method, with significant advantages, including the use of inexpensive epoxy resins, stable secondary amine bond formation between SpyDock and the resin, multiple reuse cycles, and the incorporation of a self-cleavable Mtu ∆I-CM intein to produce proteins with authentic N-termini. Mtu ∆I-CM is a pH-inducible C-terminal cleavage intein, in which C-terminal cleavage activity can be induced by a pH shift from weak alkaline to weak acid conditions [7]. It has been successfully used in several protein purification strategies for the removal of purification tags [8].
This approach provides a scalable and efficient solution for protein purification, with broad applicability in biotechnological and pharmaceutical industries. We demonstrate the success of this protocol with the purification of three model proteins: glutathione S-transferase (GST, 25.4 kDa), human growth hormone (hGH, 22.1 kDa), and the nanobody caplacizumab (27.9 kDa) [9].
Materials and reagents
Biological materials
Plasmids available upon request
1. E. coli BL21(DE3) competent cells (TransGen Biotech, catalog number: CD901-02)
2. POI (protein of interest) construct in pET30a plasmid (Merck, Novagen®, catalog number: 69909). It contains residues 1-597 with an N-terminal SpyTag002-Mtu ∆I-CM
Reagents
1. Bovine serum albumin (BSA) (Newprobe, catalog number: PB10056)
2. Premixed protein marker (Takara, catalog number: 3595A)
3. 6× protein loading buffer (TransGen Biotech, catalog number: DL101-02)
4. Sodium chloride (NaCl) (Damao Chemical Reagent Factory, CAS number: P000967)
5. Tryptone (Oxoid, catalog number: LPOO24)
6. Yeast extract (Oxoid, catalog number: LP0021)
7. Kanamycin (Newprobe, catalog number: PB1093)
8. Isopropyl-β-D-1-thiogalactopyranoside (IPTG) (Sangon Biotech, catalog number: A100487-0025)
9. Tris(2-carboxyethyl)phosphine (TCEP) (Thermo Fisher Scientific, catalog number: T2556)
10. Sodium dihydrogen phosphate (NaH2PO4) (Sangon Biotech, catalog number: A501726)
11. Disodium hydrogen phosphate (Na2HPO4) (Sangon Biotech, catalog number: A501727)
12. Potassium dihydrogen phosphate (KH2PO4) (Sangon Biotech, catalog number: A600445)
13. Imidazole (Sigma-Aldrich, catalog number: V900153)
14. Potassium chloride (KCl) (Aladdin, catalog number: P112134)
15. Sodium sulfate (Na2SO4) (Sigma-Aldrich, catalog number: 238597)
16. Ethanolamine (Aladdin, catalog number: E103810)
17. Sodium hydroxide (NaOH) pellets (Damao Chemical Reagent Factory, catalog number: P000880)
18. Hydrochloric acid (HCl) (Cologne Chemical Reagent Factory, catalog number: LC149502)
19. Acetic acid (Sangon Biotech, catalog number: A501931)
20. Tris base (Biofroxx, catalog number: 1115GR500)
21. Phosphoric acid (Aladdin, catalog number: P112024)
22. Sodium azide (NaN3) (Sigma-Aldrich, catalog number: S2002)
23. Ethylenediaminetetraacetic acid (EDTA·Na2) (Macklin, catalog number: E909976)
24. Bis-Tris (Aladdin, catalog number: B105639)
25. Guanidine hydrochloride (GdnHCl) (Aladdin, catalog number: G108676)
26. Ethanol (EtOH) (Damao Chemical Reagent Factory, CAS number: 64-17-5)
27. YoungPAGE MES SDS running buffer, powder (GenScript, catalog number: M00677)
28. SOC medium (Sangon Biotech, catalog number: B540119)
Solutions
1. LB medium (see Recipes)
2. 50 mg/mL kanamycin stock (see Recipes)
3. 1 M IPTG (see Recipes)
4. Binding buffer (see Recipes)
5. Elution buffer (see Recipes)
6. 100 mM PBS (see Recipes)
7. 1 M NaH2PO4 (see Recipes)
8. 1 M Na2HPO4 (see Recipes)
9. Coupling buffer (see Recipes)
10. 1 M ethanolamine (see Recipes)
11. 0.1 M acetate buffer (see Recipes)
12. 0.1 M Tris-HCl buffer (see Recipes)
13. Tris-phosphate (TP) buffer (see Recipes)
14. Storage buffer stock (see Recipes)
15. Wash buffer WTP (see Recipes)
16. Cleavage buffer (see Recipes)
17. Wash buffer (see Recipes)
18. 6 M GdnHCl (see Recipes)
19. 0.1 M NaOH (see Recipes)
20. 20% ethanol (see Recipes)
Recipes
1. LB medium (1,000 mL)
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| NaCl | 10 g/L | 10 g |
| Tryptone | 10 g/L | 10 g |
| Yeast extract | 5 g/L | 5 g |
| H2O | n/a | To 1,000 mL |
| Total | n/a | 1,000 mL |
Sterilize using an autoclave at 121 °C for 20 min.
2. 50 mg/mL kanamycin stock (10 mL)
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Kanamycin | 50 mg/mL | 0.5 g |
| H2O | n/a | To 10 mL |
| Total | n/a | 10 mL |
a. Sterilize by filtering with a 0.22 μm syringe filter.
b. Store in 1 mL aliquots at -20 °C.
3. 1 M IPTG (10 mL)
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| IPTG | 1 M | 2.38 g |
| H2O | n/a | To 10 mL |
| Total | n/a | 10 mL |
a. Sterilize by filtering with a 0.22 μm syringe filter.
b. Store in 1 mL aliquots at -20 °C.
4. Binding buffer (pH 7.4) (1,000 mL)
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Na2HPO4 | 15.6 mM | 2.22 g |
| KH2PO4 | 4.4 mM | 0.6 g |
| NaCl | 500 mM | 29.22 g |
| Imidazole | 30 mM | 2.04 g |
| H2O | n/a | To 1,000 mL |
| Total | n/a | 1,000 mL |
a. Adjust pH to 7.4 with 12 M HCl.
b. Filter through a 0.22 μm hydrophilic membrane.
5. Elution buffer (pH 7.4) (1,000 mL)
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Na2HPO4 | 15.6 mM | 2.22 g |
| KH2PO4 | 4.4 mM | 0.6 g |
| NaCl | 500 mM | 29.22 g |
| Imidazole | 500 mM | 34.04 g |
| H2O | n/a | To 1,000 mL |
| Total | n/a | 1,000 mL |
a. Adjust pH to 7.4 with 12 M HCl.
b. Filter through a 0.22 μm hydrophilic membrane.
6. 100 mM PBS (pH 7.0) (1,000 mL)
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Na2HPO4 | 94.7 mM | 13.44 g |
| KH2PO4 | 5.3 mM | 0.72 g |
| NaCl | 137 mM | 8 g |
| KCl | 2.7 mM | 0.2 g |
| H2O | n/a | To 1,000 mL |
| Total | n/a | 1,000 mL |
a. Adjust pH to 7.0 with 12 M HCl.
b. Filter through a 0.22 μm hydrophilic membrane.
7. 1 M NaH2PO4 (500 mL)
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| NaH2PO4 | 1 M | 59.99 g |
| H2O | n/a | To 500 mL |
| Total | n/a | 500 mL |
8. 1 M Na2HPO4 (500 mL)
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Na2HPO4 | 1 M | 70.98 g |
| H2O | n/a | To 500 mL |
| Total | n/a | 500 mL |
9. Coupling buffer (pH 10.0) (1,000 mL)
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Na2SO4 | 250 mM | 35.51 g |
| 1 M NaH2PO4 | 6.8 mM | 6.8 mL |
| 1 M Na2HPO4 | 93.2 mM | 93.2 mL |
| H2O | n/a | To 1,000 mL |
| Total | n/a | 1,000 mL |
a. Adjust pH to 8.0 with extra 1 M Na2HPO4.
b. Adjust pH from 8.0 to 10.0 with 1 M NaOH.
c. Filter through a 0.22 μm hydrophilic membrane.
10. 1 M ethanolamine (pH 8.0) (200 mL)
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Ethanolamine | 1 M | 12.1 mL |
| H2O | n/a | To 200 mL |
| Total | n/a | 200 mL |
a. Adjust pH to 8.0 with 12 M HCl.
b. Filter through a 0.22 μm hydrophilic membrane.
11. 0.1 M acetate buffer (pH 4.0) (100 mL)
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Acetic acid | 0.1 M | 571.9 μL |
| NaCl | 0.5 M | 2.922 g |
| H2O | n/a | To 100 mL |
| Total | n/a | 100 mL |
a. Adjust pH to 4.0 with 1 M NaOH.
b. Filter through a 0.22 μm hydrophilic membrane.
12. 0.1 M Tris-HCl buffer (pH 8.0) (100 mL)
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Tris base | 0.1 M | 1.2114 g |
| NaCl | 0.5 M | 2.922 g |
| H2O | n/a | To 100 mL |
| Total | n/a | 100 mL |
a. Adjust pH to 8.0 with 12 M HCl.
b. Filter through a 0.22 μm hydrophilic membrane.
13. Tris-phosphate (TP) buffer (pH 7.0) (1,000 mL)
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Phosphoric acid | 25 mM | 0.86 mL |
| H2O | n/a | To 1000 mL |
| Total | n/a | 1000 mL |
a. Adjust pH to 7.0 with Tris base.
b. Filter through a 0.22 μm hydrophilic membrane.
14. Storage buffer stock (50 mL)
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| NaN3 | 20% (w/v) | 0.01 g |
| TP buffer | n/a | To 500 mL |
| Total | n/a | 50 mL |
a. Sterilize by filtering with a 0.2 μm syringe filter.
b. Store at -20 °C.
c. Dilute 400-fold with TP buffer before use.
15. Wash buffer WTP (pH 7.0) (500 mL)
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Phosphoric acid | 25 mM | 0.86 mL |
| Imidazole | 0.5 M | 3.47 g |
| H2O | n/a | To 500 mL |
| Total | n/a | 500 mL |
a. Adjust pH to 7.0 with Tris base.
b. Filter through a 0.22 μm hydrophilic membrane.
16. Cleavage buffer (pH 6.2) (200 mL)
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Na2HPO4 | 10.2 mM | 0.288 |
| KH2PO4 | 1.8 mM | 0.048 |
| NaCl | 137 mM | 1.6 g |
| KCl | 2.7 mM | 0.04 g |
| Bis-Tris | 20 mM | 0.839 g |
| EDTA·Na2 | 2 mM | 0.1489 g |
| H2O | n/a | To 200 mL |
| Total | n/a | 200 mL |
a. Adjust pH to 6.2 with 12 M HCl.
b. Filter through a 0.22 μm hydrophilic membrane.
17. Wash buffer (pH 7) (500 mL)
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Phosphoric acid | 25 mM | 0.86 mL |
| Imidazole | 4 M | 138.94 g |
| H2O | n/a | To 500 mL |
| Total | n/a | 500 mL |
a. Adjust pH to 7 with Tris base.
b. Filter through a 0.22 μm hydrophilic membrane.
18. 6 M GdnHCl (pH 2.0) (500 mL)
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Guanidine hydrochloride | 6 M | 289.5 g |
| H2O | n/a | To 500 mL |
| Total | n/a | 500 mL |
a. Adjust pH to 2.0 with 12 M HCl.
b. Filter through a 0.22 μm hydrophilic membrane.
19. 0.1 M NaOH (500 mL)
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| NaOH | 0.1 M | 2 g |
| H2O | n/a | To 500 mL |
| Total | n/a | 500 mL |
Filter through a 0.22 μm hydrophilic membrane.
20. 20% ethanol (1,000 mL)
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Ethanol | 20% (v/v) | 200 mL |
| H2O | n/a | To 1,000 mL |
| Total | n/a | 1,000 mL |
Filter through a 0.22 μm hydrophilic membrane.
Laboratory supplies
1. HisTrapTM HP column (Cytiva, catalog number: 17524802)
2. Epoxy Activated Sepharose® 6B (Cytiva, catalog number: 17048001)
3. Empty spin columns (Biocomma, catalog number: 007400)
4. 0.2 mL PCR tubes (Axygen, catalog number: PCR-2CP-RT-C)
5. 1.5 mL microcentrifuge tubes (Axygen, catalog number: MCT-150-C)
6. 2 mL microcentrifuge tubes (Axygen, catalog number: MCT-200-C)
7. 15 mL centrifuge tubes (Corning, catalog number: 430791)
8. 50 mL centrifuge tubes (Corning, catalog number: 430829)
9. 1 L conical flask (Northglass, catalog number: HX-1121-01-11)
10. Sterile spreader (Baroque, catalog number: P003277)
11. 0.22 μm syringe filters (Pall Corporation, catalog number: 4652)
12. Syringes 20 mL, sterile, disposable (Double-dove, catalog number: HX-SLZSQ-SG-20ML/16)
13. Hydrophilic membrane filter, 0.22 μm, 47 mm, disposable (Millipore Sigma, catalog number: GSWP04700)
14. Amicon Ultra centrifugal filter units (3 K MWCO) (Millipore Sigma, catalog number: UFC900396)
15. 4%–20% YoungPAGE® Bis-Tris SDS-PAGE gel (GenScript, catalog number: M00928)
16. SnakeSkinTM dialysis tubing, 3.5 K MWCO (Thermo Fisher Scientific, catalog number: 88244)
Equipment
1. Pipetting system (Gilson, model: PIPETMAN Classic Starter Kit)
2. Shaking incubator (New Brunswick Scientific, model: Innova44)
3. Biochemical incubator (Thermo Fisher Scientific, model: IMH100-S)
4. Spectrophotometer (Youke Instrument, model: UV755B)
5. Electronic analytical balance (Mettler Toledo, model: MS204TS)
6. Autoclave (Hirayama, model: HVE-50)
7. -80 °C freezer (Thermo Fisher Scientific, model: 995)
8. Clean bench (Donglian, model: DL-CJ-2NDII)
9. Ultrasonic crasher (Scientz, model: JY92-II)
10. Centrifuge for 15 and 50 mL tubes (Eppendorf, model: 5810R)
11. Centrifuge for 1 and 2 mL tubes (Eppendorf, model: 5425R)
12. Electrophoresis equipment (Bio-Rad, model: Mini-PROTEAN® Tetra Cell Systems)
13. pH meter (Mettler Toledo, model: FE28-standard)
14. Mini rotator (Gilson, model: Roto-MiNi Plus)
15. AKTA Purifier chromatography system (GE Healthcare, model: AKTA Purifier 10)
16. Protein staining instrument (GenScript, model: eStain L1)
17. Vacuum filtration system (Ilmvac GmbH, model: 120788)
18. Dry bath incubator (Coyote, model: H2O3-100C)
19. Magnetic stirrer (IKA, model: RTC-basic)
Software and datasets
1. SnapGene® (SnapGene, https://www.snapgene.com/)
2. ImageJ (National Institutes of Health DNA, https://imagej.net/ij/)
3. Prism (GraphPad, https://www.graphpad.com/)
4. Compute pI/Mw tool (https://web.expasy.org/compute_pi/)
5. Unicorn for AKTA purifier (GE Healthcare, https://www.cytivalifesciences.com/en/us/shop/chromatography/software/unicorn-7-p-05649)
Procedure
Note: Perform most procedures at room temperature (25 °C), except for steps where specific temperatures are indicated.
A. Protein expression
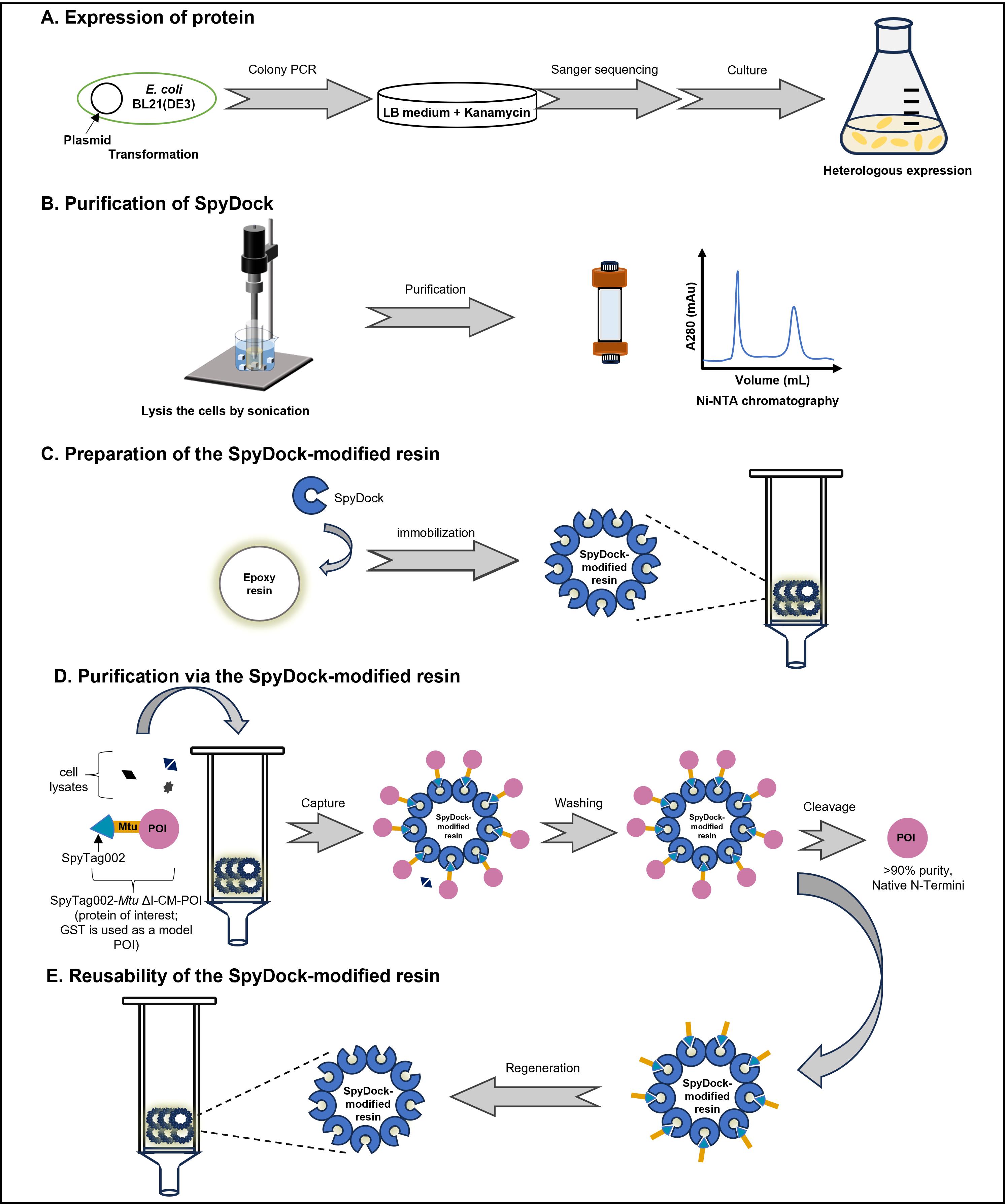
1. Transform E. coli BL21(DE3) bacterial strain with the kanamycin-resistant pET30a-6×His-GS linker-SpyDock plasmid for SpyDock production (Figure 1A) or with the kanamycin-resistant pET30a-SpyTag002-Mtu ∆I-CM-POI plasmid for protein of interest (POI) production. Glutathione S-transferase (GST) is used as a model protein (SpyTag002-Mtu ∆I-CM-GST) in this protocol (Figure 1B). Use the standard heat-shock method for transformation:
a. Add 1–50 ng of plasmid DNA to 100 μL of chemically competent cells. Mix by gentle tapping and place on ice.
b. Incubate the cells on ice for 30 min.
c. Transfer the cells to a 42 °C biochemical incubator for exactly 90 s.
d. Transfer the cells to ice for 2 min.
e. Add 900 μL of SOC medium to the cells.
f. Incubate the cells on a shaking incubator (220 rpm) at 37 °C for 45 min.
g. Pipette 200 μL of transformed cell suspension onto an LB agar plate with kanamycin and spread it using a sterile spreader.
h. Incubate the plate overnight at 37 °C.
i. Perform colony PCR and Sanger sequencing to select an E. coli colony transformed with a plasmid vector containing the insert fragment.
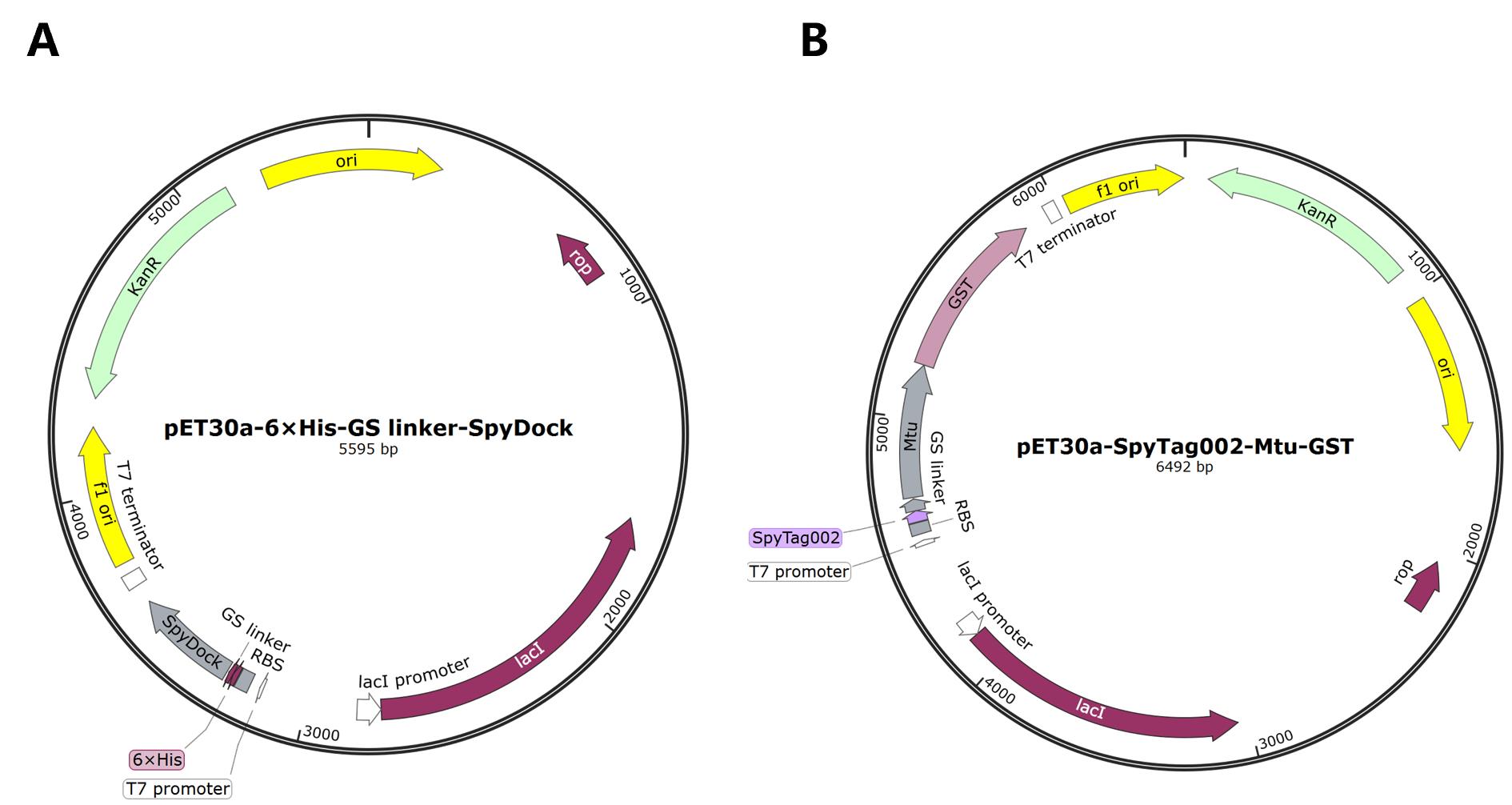
Figure 1. Plasmid maps. (A) pET30a-6×His-GS linker-SpyDock. (B) pET30a-SpyTag002-Mtu ∆I-CM-GST.
2. Starter culture: Inoculate 5 mL of LB medium containing 50 μg/mL kanamycin with a single colony of bacteria in a 50 mL tube; grow overnight (~12–16 h) at 37 °C with shaking at 220 rpm.
3. Main culture: Dilute the saturated overnight culture 1:50 in fresh LB medium containing 50 μg/mL kanamycin in a conical flask and incubate at 37 °C with shaking at 220 rpm until the optical density at 600 nm (OD600) reaches 0.4–0.6 (~2 h).
4. After ~2 h, add IPTG to a final concentration of 0.2 mM and let the bacteria grow at 18 °C, 220 rpm for 20–24 h.
5. After 20–24 h, pipette 1 mL of the culture to measure the final OD600 and then transfer the remaining culture into 50 mL centrifuge tubes.
6. Pellet bacteria by centrifugation at 8,200× g for 15 min at 4 °C. Store the cell pellet at -80 °C until use.
B. SpyDock purification
1. Thaw a bacterial cell pellet on ice. Thoroughly resuspend the pellet in ice-cold binding buffer to a final OD600 of 50 by pipetting up and down using a 1 mL micropipette tip.
Note: Calculate the resuspension volume based on the initial culture volume and the measured OD600 value (as described in step A5) to concentrate the bacterial cells to an OD600 of 50 for further purification. For example, if the cells are harvested at an OD600 of 5, resuspend the cell pellet from each 10 mL of culture in 1 mL of binding buffer.
2. Disrupt the cell suspension on ice using an ultrasonic crasher for 30 min with the following settings: 3 s ON, 3 s OFF, power = 195 watts, amplitude probe = 6 mm. Check if the suspension has become less viscous and slightly darker.
3. Centrifuge the lysate at 18,500× g for 20 min at 4 °C to pellet insoluble material and debris and transfer the supernatant (cleared lysate) to a new pre-chilled centrifuge tube on ice.
4. Perform Ni-NTA affinity purification using a fast protein liquid chromatographic system (AKTA purifier) with a 5 mL HisTrapTM HP column connected in series. Equilibrate a HisTrapTM HP column with 10 column volumes (CVs) of binding buffer at a 5 mL/min flow rate. Filter the supernatant using a 0.22 μm syringe filter and load it onto the pre-equilibrated column at a 1 mL/min flow rate.
5. After loading, wash the column with binding buffer until the absorbance at 280 nm reaches a steady baseline.
6. Elute the SpyDock protein using a linear gradient from 0% to 100% elution buffer for 30 CVs. Collect elution fractions (~12.5 mL per fraction) in 50 mL centrifuge tubes and store them at 4 °C.
7. Wash the column with elution buffer to remove any residual bound protein. Pass 25 mL of ddH2O through the column and store the Ni2+-NTA column in 20% ethanol.
8. Analyze the elution fractions on 4%–20% YoungPAGE® Bis-Tris SDS-PAGE gel (Figure 2).
a. Mix an aliquot of collected target protein with protein loading buffer in a 5:1 volume ratio.
b. Heat the protein samples at 98 °C for 10 min.
c. Run the protein samples on SDS-PAGE gels.
d. Visualize the protein in SDS-PAGE gels with eStain L1 protein staining system.
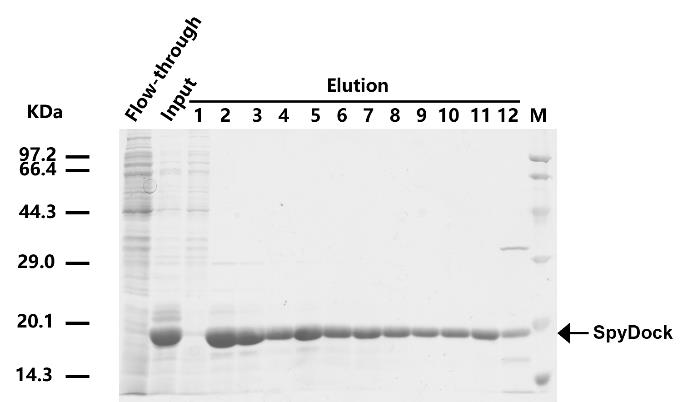
Figure 2. SDS-PAGE for the SpyDock purified by affinity chromatography
9. Choose fractions with high SpyDock protein purity (above 90%, fractions 2–11 as shown in Figure 2) and concentrate them using Millipore centrifugal filters (3 kDa cutoff) at 3,220× g in an Eppendorf benchtop centrifuge 5810 R at 4 °C.
10. Carefully transfer the concentrated protein into a dialysis bag (3.5 kDa MWCO). Seal the bag tightly and place it in a beaker containing 2 L of coupling buffer. Dialyze against the coupling buffer at 4 °C overnight with gentle stirring using a magnetic stirrer.
Note: A yield of 50–100 mg purified SpyDock per liter of culture in a shake flask scale was collected in the form of protein solution.
C. Preparation of the SpyDock-modified resin
1. Preparation of epoxy-activated Sepharose 6B
a. Weigh out the required amount of freeze-dried epoxy-activated Sepharose 6B powder (1 g of freeze-dried powder produces approximately 3.5 mL of final volume of medium) and suspend it in double-distilled water (ddH2O).
b. The medium swells immediately and should be washed for 1 h on a sintered glass filter. Use approximately 200 mL of ddH2O per gram of freeze-dried powder, added in several aliquots.
c. Wash the swollen resin five times with coupling buffer. For small-scale preparations, use 500 μL of coupling buffer for 250 μL of resin.
2. Prepare the SpyDock solution by adding TCEP to a final concentration of 1 mM. Incubate the solution at 25 °C for 30 min to reduce the SpyDock.
Note: Collect the purified SpyDock protein obtained per preparation as a protein solution. For small-scale preparation, dissolve 25 mg of SpyDock in 250 μL of coupling buffer containing 1 mM TCEP.
3. Add 100 mg of reduced SpyDock per milliliter of prepared epoxy-activated Sepharose 6B. Mix the resin slurry at 25 °C, 14 rpm, for 12 h.
Note: For small-scale preparation, add 25 mg of SpyDock for 250 μL of resin in this protocol. Adjust volumes and amounts proportionally based on experimental needs. The amount of SpyDock is measured against standard albumin in the case of a solution.
4. Collect the resulting resin by centrifugation at 100× g for 1 min at 4 °C. Wash the resin five times with 2 CVs of coupling buffer to remove unbound SpyDock.
5. Incubate the resin in 2 CVs of 1 M ethanolamine solution at 37 °C for 12 h to block the excess remaining epoxy groups.
6. After 12 h, wash the resulting resin with 2 CVs of 0.1 M acetate buffer, followed by 2 CVs of 0.1 M Tris-HCl buffer, for three cycles. Store the prepared SpyDock-modified resin (Figure 3) in storage buffer at 4 °C until use.
Note: SpyDock-modified resin remains functional after being stored in 20% ethanol at 4 °C for 3–6 months.
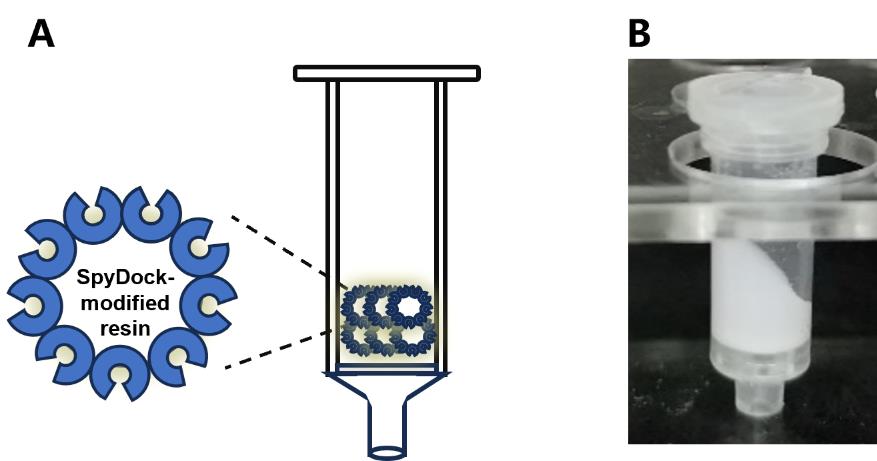
Figure 3. Diagram of SpyDock-modified resin assembly. (A) Schematic and (B) photo depicting column apparatus. Figure from Yang et al. [8]. See Section C for details.
D. Purification of GST via the SpyDock-modified resin
1. Thoroughly resuspend the bacterial cell pellet [SpyTag002-Mtu ∆I-CM-GST (glutathione S-transferase)] and disrupt the cell suspension on ice using an ultrasonic crasher for 30 min, as described in section B.
2. Mix 250 μL of the SpyDock-modified resin with 400 μL of the E. coli cell lysates containing SpyTag002-Mtu ∆I-CM-GST (glutathione S-transferase) on a rotator at 25 °C, 14 rpm, for 2 h to allow for efficient binding.
Note: The SpyTag-fused target protein is specifically captured by the SpyDock-modified resin through specific Spy chemistry. Structures are shown from SpyTag/SpyCatcher (PDB ID 4MLI).
3. After 2 h, pellet the resin by centrifugation at 100× g for 1 min at 4 °C. Transfer the supernatant (flowthrough), which contains the unbound protein, to a new microcentrifuge tube (e.g., 1.5 mL tube) for further analysis if needed.
4. Wash the resin five times at room temperature with 500 μL of WTP buffer to remove nonspecifically bound proteins.
5. Resuspend the resin in 400 μL of cleavage buffer and incubate at 37 °C for 3 h to induce the cleavage of the captured SpyTag002-Mtu ∆I-CM-GST, releasing the GST.
6. After 3 h, pellet the resin by centrifugation at 100× g for 1 min at 4 °C. Transfer the suspension containing the GST to a new microcentrifuge tube.
7. Wash the resin five times at room temperature with 500 μL of TP buffer.
8. Analyze the collected target protein on 4%–20% SDS-PAGE gel as described in section B (Figure 4).
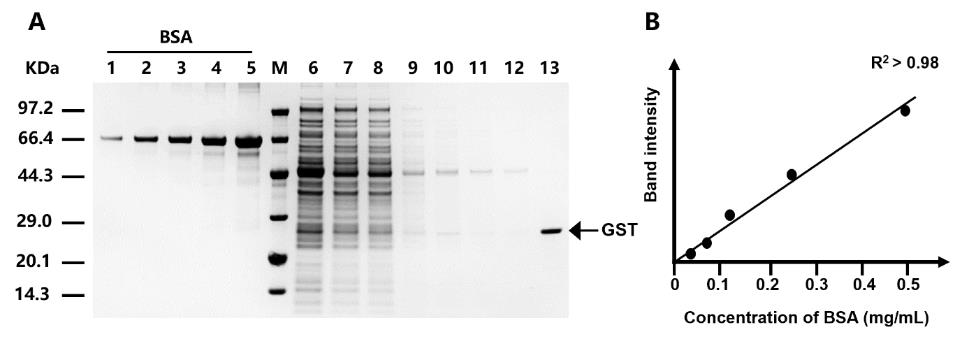
Figure 4. SDS-PAGE analysis of purification steps using SpyDock-modified resin. The purification of glutathione S-transferase (GST) is shown as an example to illustrate the purification process. GST derived from Smith et al. [10]. Lanes 1–5: BSA standards with concentrations of 0.03125, 0.0625, 0.125, 0.25, and 0.5 mg/mL, respectively; Lane 6: protein sample prior to purification; Lane 7: unbound proteins; Lanes 8–12: removal of nonspecific bound proteins by washing; Lane 13: purified target protein through intein-mediated cleavage. Figure from Yang et al. [9].
E. Reusability of the SpyDock-modified resin
1. After elution, wash the resin three times at room temperature with 1.5 CVs of wash buffer, followed by an additional wash with 500 μL of TP buffer.
2. Wash the resin three times at room temperature with 500 μL of 6 M GdnHCl, followed by an additional wash with 500 μL of TP buffer.
3. Wash the resin three times at room temperature with 500 μL of 0.1 M NaOH. Rinse the resin twice with 500 μL of TP buffer.
4. Store the SpyDock-modified resin in 20% ethanol at 4 °C.
Data analysis
To assess the purity and amounts of the proteins, run the samples on an SDS-PAGE gel. Determine protein compositions and amounts densitometrically using ImageJ software (NIH, USA), with BSA as the standard (Figure 5). Prepare a series of BSA standards at known concentrations (0.5, 0.25, 0.125, 0.0625, and 0.03125 mg/mL) and load them alongside the protein samples. After electrophoresis, stain the gel using the eStain L1 protein staining system. Measure band intensities with ImageJ and generate a standard curve by plotting known protein concentrations (x-axis) against their corresponding band intensities (y-axis). Estimate protein concentrations of the target protein (GST, shown as an example in Figure 4) by plotting the band intensity of the target protein (in the y-axis) and determining the intersection point with the BSA standard curve. Then, find the concentration associated with that particular point (in the x-axis).
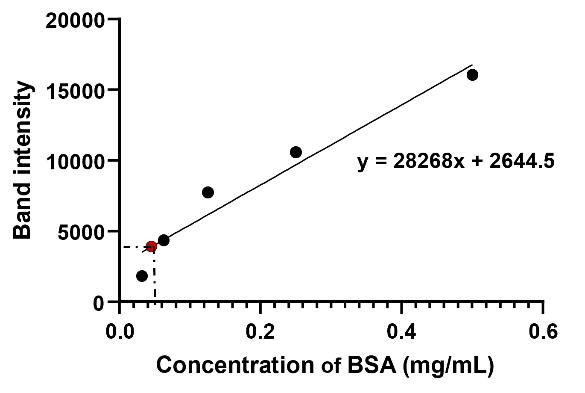
Figure 5. BSA standard curve. The black dots represent BSA band intensities at known concentrations (0.03125, 0.0625, 0.125, 0.25, and 0.5 mg/mL), which are used to generate the standard curve. The red dot represents the band intensity of the target protein (GST). This BSA curve and equation will be used to calculate GST concentration.
Validation of protocol
This protocol or parts of it has been used and validated in the following research article:
• Yang et al. [9]. A Spy chemistry-based method for purification of proteins with authentic N-termini. Catalysts. (Figures 2 and 3)
General notes and troubleshooting
General notes
1. This protocol describes cell lysis using a sonicator. For larger-scale applications, a high-pressure homogenizer can serve as an alternative. Additionally, a simple setup with a peristaltic pump may replace the AKTA purifier for HisTrapTM HP column purification.
2. SpyDock with a purity of over 90% is recommended for efficient immobilization onto epoxy resin. To prevent protein degradation and maintain high immobilization efficiency, prepare SpyDock freshly and reduce disulfide-bonded SpyDock to its monomeric form using TCEP before use.
3. This protocol is broadly applicable for purifying other proteins, provided they are expressed in a soluble form in E. coli or other expression systems.
4. To minimize protein aggregation during concentration, it is advisable to replace the tubing with a new one if aggregation occurs due to excessively high protein concentrations.
5. Bacterial pellets can be stored at -80 °C for up to 6 months.
Troubleshooting
Problem 1: Low immobilization efficiency of SpyDock.
Possible causes: The functional activity of SpyDock might decline due to prolonged storage. Additionally, the epoxy groups on the resin may become partially inactivated after extended storage.
Solution: Use freshly prepared SpyDock and replace the epoxy resin with a new batch to regain optimal immobilization efficiency.
Problem 2: Low cleavage yield of the target protein.
Possible cause: Incomplete cleavage of the target protein.
Solution: Extend the cleavage time at 37 °C or perform the cleavage step at 25 °C overnight to ensure complete processing.
Acknowledgments
Conceptualization, Z.L. (Zhanglin Lin) and X.Y(Xiaofeng Yang); Investigation, B.C. (Binrui Chen), Z.L. (Zisha Lao), and Y.X. (Ya Xiang); Writing—Original Draft, Y.X. (Ya Xiang); Writing—Review & Editing, X.Y(Xiaofeng Yang) and Z.L. (Zhanglin Lin); Funding acquisition, X.Y. (Xiaofeng Yang); Supervision, X.Y. (Xiaofeng Yang) and Z.L. (Zhanglin Lin).
This study was supported by the National Key Research and Development Program of China (2022YFC2104800).
The protocols described here were adapted from previously reported studies by the authors [9].
References
- Łącki, K. M. and Riske, F. J. (2020). Affinity Chromatography: An Enabling Technology for Large‐Scale Bioprocessing. Biotechnol J. 15(1): e201800397. https://doi.org/10.1002/biot.201800397
- Young, C. L., Britton, Z. T. and Robinson, A. S. (2012). Recombinant protein expression and purification: A comprehensive review of affinity tags and microbial applications. Biotechnol J. 7(5): 620–634. https://doi.org/10.1002/biot.201100155
- Khairil Anuar, I. N. A., Banerjee, A., Keeble, A. H., Carella, A., Nikov, G. I. and Howarth, M. (2019). Spy&Go purification of SpyTag-proteins using pseudo-SpyCatcher to access an oligomerization toolbox. Nat Commun. 10(1): 1734. https://doi.org/10.1038/s41467-019-09678-w
- Mahmoudi Gomari, M., Saraygord-Afshari, N., Farsimadan, M., Rostami, N., Aghamiri, S. and Farajollahi, M. M. (2020). Opportunities and challenges of the tag-assisted protein purification techniques: Applications in the pharmaceutical industry. Biotechnol Adv. 45: 107653. https://doi.org/10.1016/j.biotechadv.2020.107653
- Zakeri, B., Fierer, J. O., Celik, E., Chittock, E. C., Schwarz-Linek, U., Moy, V. T. and Howarth, M. (2012). Peptide tag forming a rapid covalent bond to a protein, through engineering a bacterial adhesin. Proc Natl Acad Sci USA. 109(12): e1115485109. https://doi.org/10.1073/pnas.1115485109
- Yan, H. and Chen, F. (2022). Recent Progress in Solid‐Phase Total Synthesis of Naturally Occurring Small Peptides. Adv Synth Catal. 364(12): 1934–1961. https://doi.org/10.1002/adsc.202200079
- Wood, D. W., Wu, W., Belfort, G., Derbyshire, V. and Belfort, M. (1999). A genetic system yields self-cleaving inteins for bioseparations. Nat Biotechnol. 17(9): 889–892. https://doi.org/10.1038/12879
- Fong, B. A., Wu, W. Y. and Wood, D. W. (2010). The potential role of self-cleaving purification tags in commercial-scale processes. Trends Biotechnol. 28(5): 272–279. https://doi.org/10.1016/j.tibtech.2010.02.003
- Yang, X., Chen, B., Lao, Z., Xiang, Y. and Lin, Z. (2024). A Spy Chemistry-Based Method for Purification of Proteins with Authentic N-Termini. Catalysts. 14(9): 651. https://doi.org/10.3390/catal14090651
- Smith, D. B., Davern, K. M., Board, P. G., Tiu, W. U., Garcia, E. G. and Mitchell, G. F. (1986). Mr 26,000 antigen of Schistosoma japonicum recognized by resistant WEHI 129/J mice is a parasite glutathione S-transferase. Proc Natl Acad Sci USA. 83(22): 8703–8707. https://doi.org/10.1073/pnas.83.22.8703
Article Information
Publication history
Received: Jan 6, 2025
Accepted: Mar 11, 2025
Available online: Mar 24, 2025
Published: Apr 20, 2025
Copyright
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Yang, X., Lin, Z., Xiang, Y., Chen, B. and Lao, Z. (2025). A Robust and Easy Protein Purification Method Using SpyDock-Modified Resin. Bio-protocol 15(8): e5270. DOI: 10.21769/BioProtoc.5270.
Category
Biochemistry > Protein > Isolation and purification
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









