- Submit a Protocol
- Receive Our Alerts
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Microbial Biofilm Detection and Differentiation by Dual Staining Using Maneval’s Stain
(§ Technical contact) Published: Vol 15, Iss 5, Mar 5, 2025 DOI: 10.21769/BioProtoc.5228 Views: 742
Reviewed by: Emilia KrypotouRubikah VimonishAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
Imaging Assays to Detect DNA Damage in Trypanosome Parasites Using γH2A
Rajiv S. Jumani [...] Srinivasa P. S. Rao
Jul 5, 2024 782 Views
Genetic Tagging and Imaging of Proteins with iFAST in Candida albicans
Jonas Devos [...] Wouter Van Genechten
Oct 5, 2024 514 Views
Synchronized Visualization and Analysis of Intracellular Trafficking and Maturation of Orthoflavivirus Subviral Particles
Kotaro Ishida and Eiji Morita
May 20, 2025 686 Views
Abstract
Microbial biofilms are structured communities of microorganisms embedded in a self-produced extracellular matrix, adhering to surfaces. These biofilms enhance bacterial resistance to antibiotics, immune responses, and environmental stress. Current microscopy techniques, such as scanning electron microscopy (SEM), confocal laser scanning microscopy (CLSM), and fluorescence microscopy, are commonly used to visualize and differentiate biofilms. However, their high cost and complexity often render them impractical. In contrast, simpler methods like crystal violet and Congo red staining are limited in distinguishing bacterial cells from the biofilm matrix. This study introduces a cost-effective, dual-staining method using Maneval’s stain to visualize and differentiate microbial biofilms. It requires only basic equipment and minimal reagents, making it ideal for routine use in clinical diagnosis and microbial research.
Key features
• This dual-staining method differentiates bacterial cells, biofilm matrix, and capsules in a single stain.
• This method applies to both bacterial and fungal biofilm.
• This method requires no specialized training or equipment, with the entire process completed within 30–45 min.
• Stained slides can be stored for extended periods (months) without degradation.
Keywords: Bacterial biofilmGraphical overview
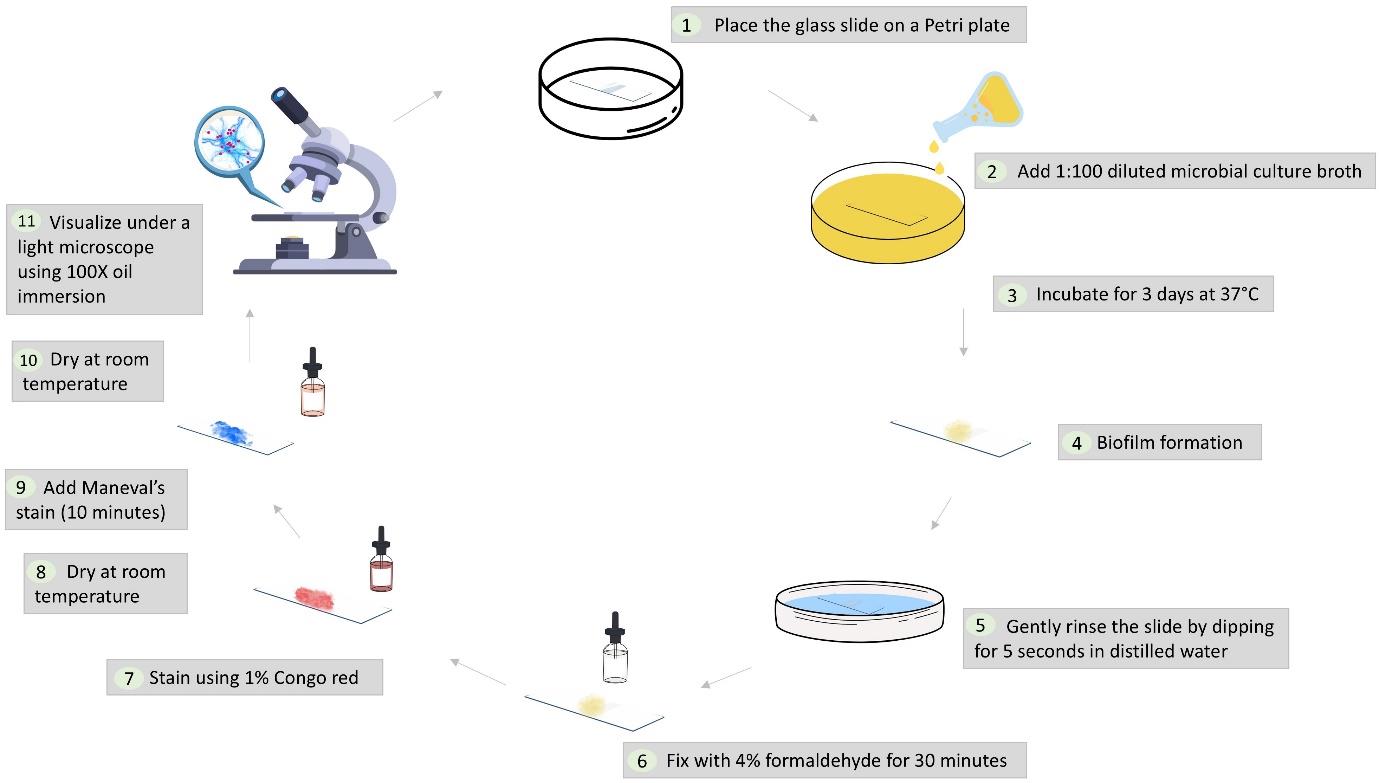
Workflow illustrating the dual-staining protocol for microbial biofilms using Maneval’s stain
Background
The discovery of biofilms dates back to the Dutch microscopist Antonie van Leeuwenhoek, who first observed them on tooth surfaces [1]. They were later studied by J.W. Costerton, who elucidated their role in persistent infections and antibiotic resistance [2]. These microbial biofilms are organized communities of microorganisms that adhere to surfaces and are encased in an extracellular matrix (ECM). The biofilm life cycle involves attachment, irreversible adhesion, maturation into microcolonies, and dispersion. The ECM, composed of polysaccharides, proteins, DNA, and lipids, acts as a protective shield, making biofilms highly resistant to antimicrobial treatments and immune responses [3].
Understanding biofilms is crucial for developing effective detection methods and treatments. Biofilm detection is crucial in clinical diagnosis, as biofilms contribute to antibiotic resistance and can lead to chronic conditions [4]. In the food industry, detecting biofilms on processing equipment is vital for preventing contamination and spoilage [5]. Monitoring biofilm growth in industrial water systems, cooling towers, and pipelines is essential to maintain operational efficiency [6]. Biofilm detection is important for studying microbial communities in natural environments, such as streams, soil, and marine ecosystems [7]. Moreover, pharmaceutical companies rely on biofilm detection methods to develop and assess the effectiveness of antibiofilm treatments [8].
Advanced microscopy techniques, such as scanning electron microscopy (SEM), confocal laser scanning microscopy (CLSM), and fluorescence microscopy, are commonly used to visualize and differentiate biofilms. However, these methods are not feasible in all laboratories because of their high cost and complexity [9]. In contrast, simpler methods like crystal violet and Congo red staining fail to distinguish bacterial cells from the biofilm matrix [10].
This study introduces a novel dual-staining method combining Maneval’s stain and Congo red stain to visualize and differentiate microbial biofilms. Maneval’s stain, initially developed by Edmond Maneval in 1941 for visualizing bacterial capsules, has been adapted in this study for biofilm detection [11]. This simple, cost-effective method requires only basic equipment and minimal reagents, making it suitable for routine laboratory use. The dual-staining method effectively differentiates bacterial cells and biofilm matrix, displaying a distinctive blue polysaccharide layer surrounding the magenta-red bacterial cells. Congo red interacts with the hydrophobic regions of polysaccharides through hydrogen bonds [12], initially staining them red. Upon adding Maneval’s stain, the pH becomes acidic, causing a shift to blue due to the protonation of the azo group [13]. Simultaneously, acid fuchsin binds to the negatively charged bacterial surfaces, creating a magenta-red coloration.
We applied this technique to various microbial species, including Gram-positive bacteria with a thick peptidoglycan layer (Staphylococcus aureus and Enterococcus faecalis), Gram-negative bacteria distinguished by their outer membrane (Escherichia coli and Pseudomonas aeruginosa), and fungi (Candida albicans), which differ significantly in the presence of chitin, glucan, and mannoproteins. While this method does not provide the detailed structural resolution of high-magnification techniques like SEM, it offers a practical and accessible alternative for distinguishing biofilm matrix and bacterial cells.
Materials and reagents
Biological materials
1. Microbial culture (any bacterial or fungal culture including clinical isolates or standard strains such as ATCC)
Reagents
1. 1% Congo red (HIMEDIA Laboratories, catalog number: GRM927)
2. Maneval’s stain (Carolina Biological Supply, catalog number: 873785 or prepared in-house, see Recipe 2)
3. 4% Formaldehyde
Caution: Formaldehyde is a hazardous chemical and should be handled in a well-ventilated area with appropriate personal protective equipment (PPE), including gloves, lab coat, and safety goggles.
4. Nutrient broth (HIMEDIA Laboratories, catalog number: M002) or any basic broth (e.g., LB broth, TSB broth, BHI broth)
Solutions
1. 1% Congo red solution (see Recipes)
2. Maneval’s stain (see Recipes)
Recipes
1. 1% Congo red solution
| Reagent | Quantity or Volume |
|---|---|
| Congo red | 1 g |
| Distilled water | 100 mL |
2. Maneval’s stain [14]
| Reagent | Quantity or Volume |
|---|---|
| Fuchsin | 0.05 g |
| Ferric chloride | 3.0 g |
| Acetic acid | 5 mL |
| Phenol | 3.9 mL |
| Distilled water | 95 mL |
Laboratory supplies
1. Glass slides [Blue Star (PIC1), 1.35 mm thick, 75 mm× 25 mm]
2. Petri plates (SRL Pvt. Ltd., polystyrene 90 mm × 15 mm, catalog number: 90226)
3. Droppers (Plastic Labware India, catalog number: PPPD101)
4. Staining rack (Advance Scientific & Surgical Co., catalog number: ADV-188)
5. Microscope oil (cedarwood oil) (SRL Pvt. Ltd., catalog number: 31018)
6. Gloves (Microgene Diagnostic Systems Pvt. Ltd.)
Equipment
1. Light microscope (Radical Scientific Equipment Pvt. Ltd., model: (RXL-5 Binocular Research Microscope)
2. Incubator (set to 37 °C)
Procedure
A. Preparation of biofilm on slides
1. Prepare a 1:100 dilution of 0.5 McFarland-adjusted microbial culture using sterilized nutrient broth. Place a sterilized glass slide in a 90 mm × 15 mm polystyrene Petri dish and fill it with the diluted culture broth to ensure complete submersion of the slide. To prepare a polymicrobial culture, mix equal volumes of microbial suspensions. For example, combine 500 μL each of Staphylococcus aureus and Pseudomonas aeruginosa (1:100 dilution) to prepare a total volume of 1 mL. Ensure all microbial cultures are in the same growth phase.
Note: Do not use a high inoculum of microbial culture; see Troubleshooting 7.
2. Incubate the setup at 37 °C in undisturbed conditions for 3 days to allow biofilm formation.
Note: The culture media is not changed during the incubation to avoid contamination.
B. Rinsing of slides
1. Gently rinse the slide by dipping it in distilled water for 5 s.
Note: Avoid excessive force to prevent disturbing the biofilm structure; see Troubleshooting 1.
C. Fixation of biofilm
1. Fix the biofilm by immersing the slide in 4% formaldehyde for 15–30 min at room temperature.
2. Allow the slide to air dry completely after fixation (5–10 min).
Note: Avoid air drying the slides for extended periods to prevent crack formation in the biofilm structure; see Troubleshooting 3.
D. Staining with Congo red
1. Apply 1% Congo red stain to the slide, ensuring even biofilm coverage.
2. Remove excess stain by gently tilting the slide to allow runoff.
3. Air dry the slide for 5–10 min.
Note: Do not wash the slides; see Troubleshooting 9.
E. Staining with Maneval’s stain
1. Apply Maneval’s stain to the slide, fully covering the biofilm.
2. Incubate the slide with the stain for 10 min at room temperature.
Note: Follow the incubation times for proper staining; see Troubleshooting 8.
3. After incubation, remove excess stain (by tilting the slides) and air dry the slide for 5 min.
F. Microscopic visualization of biofilm
1. Observe the stained biofilm under a light microscope using 100× oil immersion.
2. Capture representative images for documentation and analysis.
Data analysis
1. Image capture
Biofilm images are captured under a light microscope using a 100× oil immersion objective to visualize the biofilm matrix and bacterial cells.
2. Color interpretation
a. The blue color in the stained images corresponds to the biofilm matrix stained with Congo red (Figure 1).
Note: If the biofilm matrix is not properly stained, see Troubleshooting 9.
b. The magenta-red color indicates bacterial or fungal cells, visualized using Maneval's stain (Figure 1).
Note: If bacterial or fungal cells appear dull or faded, see Troubleshooting 8.
c. Halo formation: A halo around bacteria/yeast, which indicates the presence of a capsule, is assessed. The capsule appears as a clear zone surrounding the bacterial cell (Figure 1B and C).
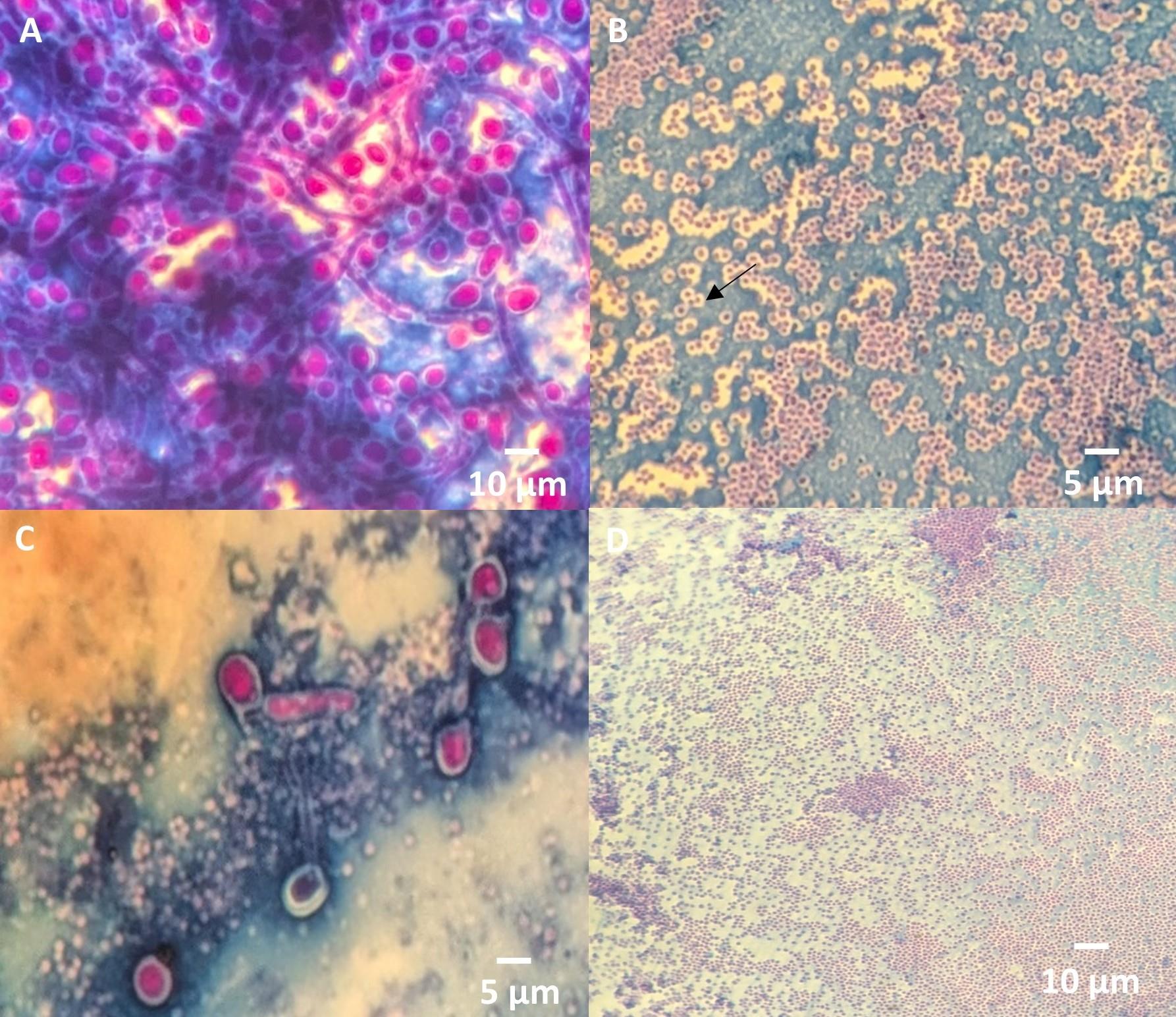
Figure 1. Stained images from the dual-staining protocol using Maneval’s stain. (A) The biofilm matrix appears blue, and bacteria/yeast cells are magenta red colored. (B) Halo formation around S. aureus indicates a capsule as a clear zone surrounding the cell. (C) Halo formation around C. albicans highlights capsule presence. (D) Negative control of biofilm (RN6390 strain).
3. Biofilm structural stages
a. Early biofilm formation: Biofilms begin with cobweb-like structures. These structures are thin, disorganized, and lack a clear shape, often forming at the interface between bacteria and the surface [15] (Figure 2A).
b. Intermediate stage: As the biofilm matures, it transitions to a clustered stage, where biofilm begins to form thicker layers with more defined structures. Bacteria accumulate and form small clusters within the biofilm matrix [15] (Figure 2B).
c. Mature biofilm: In the final stage, the biofilm develops a more honeycomb-like structure, characterized by well-defined channels and interstitial spaces that allow nutrient flow. At this stage, the cells within the biofilm may also exhibit more resistance to antimicrobial agents [16] (Figure 2C).
Note: Focus on the corners of the biofilm; see General note 8.
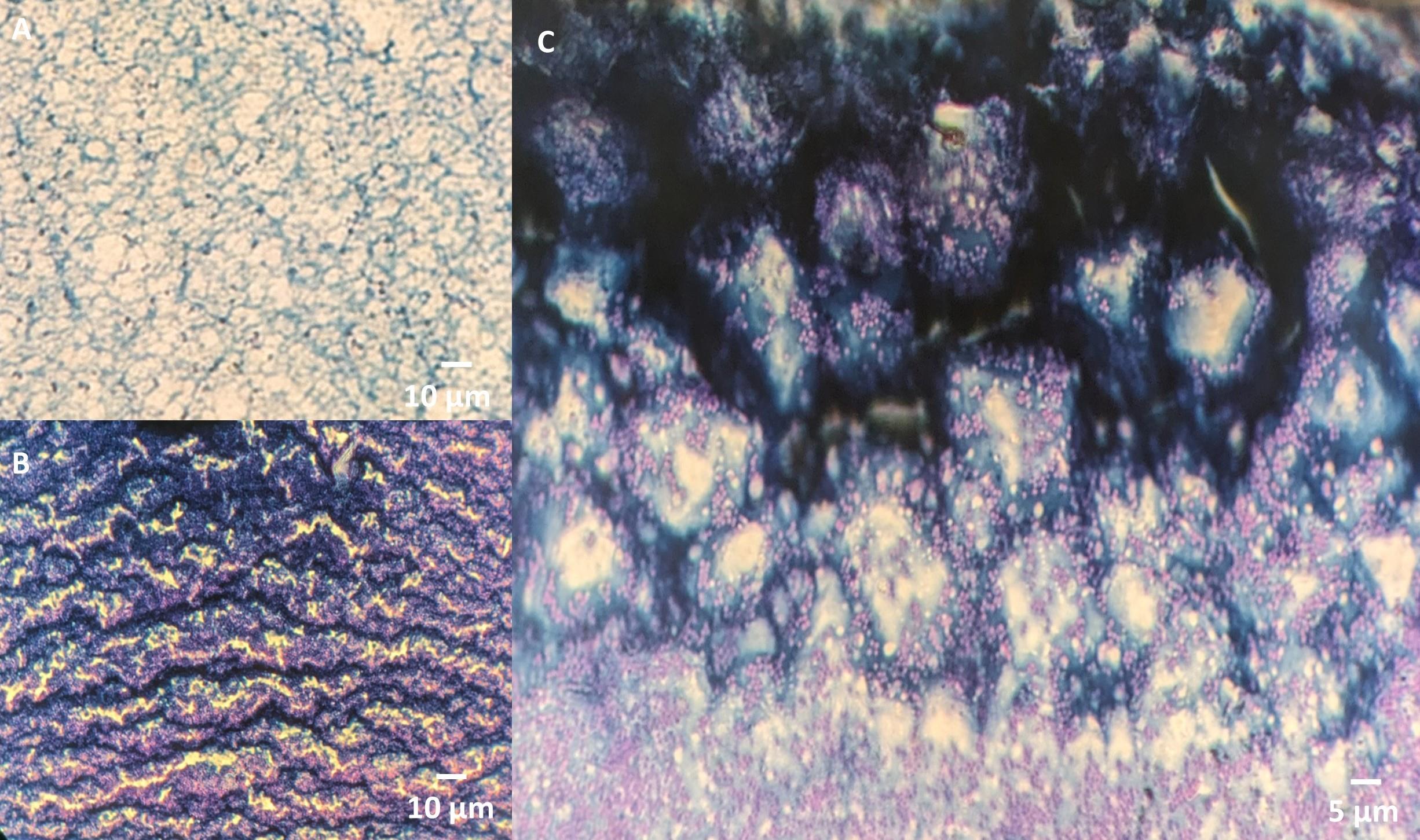
Figure 2. Biofilm structural stages are visualized using the dual-staining method. (A) Early biofilm formation shows disorganized, cobweb-like structures. (B) The intermediate stage exhibits thicker layers with small bacterial clusters within the biofilm matrix. (C) Mature biofilm displays a honeycomb-like structure with defined channels and interstitial spaces, supporting nutrient flow and increased antimicrobial resistance.
Validation of protocol
1. Evaluation of the dual-staining method on diverse microbial species: We tested this method on various organisms representing different microbial classes to validate this method. The procedure was applied to Gram-positive bacteria with a thick peptidoglycan layer, such as Staphylococcus aureus and Enterococcus faecalis, Gram-negative bacteria with an outer membrane, such as Escherichia coli and Pseudomonas aeruginosa, and fungi which differ significantly in the presence of chitin, glucan, and mannoproteins in their cell wall (Candida albicans). This diverse selection of microorganisms ensured the robustness and applicability of the method across various microbial groups (Figure 3).
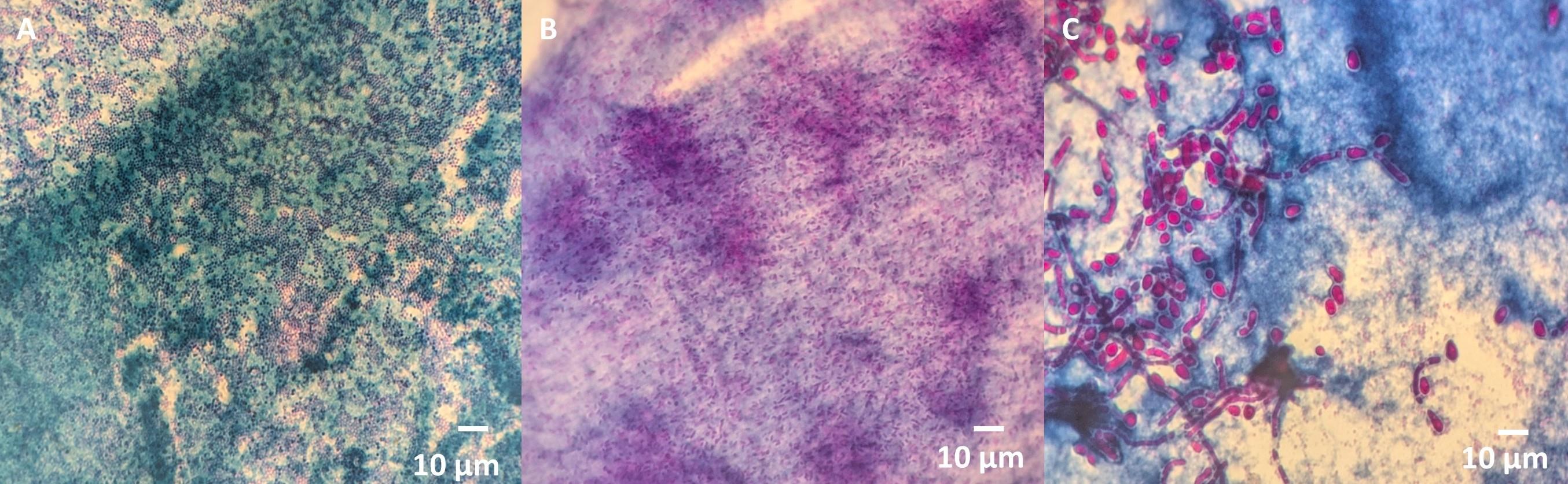
Figure 3. Validation of the dual-staining method on diverse microbial species. (A) Gram-positive bacteria (Staphylococcus aureus). (B) Gram-negative bacteria (Pseudomonas aeruginosa). (C) Fungi (Candida albicans).
2. Controls: Negative controls included media without inoculum and non-biofilm-producing bacterial strains. Biofilm-forming bacterial strains were used as a positive control [17].
3. Replicates: Each experiment was conducted in triplicate to ensure reliability.
4. Comparison with the gold standard method: Biofilm presence or absence was compared to the microtiter plate assay. The biofilm detection results were assessed using Cohen’s kappa test to evaluate agreement between the two methods. A 2 × 2 confusion matrix was created, incorporating true positive, false positive, false negative, and true negative values. Additionally, a receiver operating characteristic (ROC) curve was generated, and the area under the curve (AUC) was calculated to assess the performance of the dual-staining method. K-Fold cross-validation was conducted using 5-fold for further validation. For details, refer to the original article [17].
5. Comparison with other microscopic biofilm detection methods: The dual-staining method was further validated by comparing it with other established microscopic biofilm detection techniques, including crystal violet staining, Congo red staining, scanning electron microscopy (SEM), confocal laser scanning microscopy (CLSM), and fluorescence microscopy. Refer to the original article for detailed comparisons [17].
This protocol has been used and validated in the following research article:
Nirmala, B. et al. [17] A novel dual-staining method for cost-effective visualization and differentiation of microbial biofilms. Scientific Reports (Figures 4–6).
General notes and troubleshooting
General notes
1. Sample type: This protocol is optimized for bacterial biofilms formed on glass slides. It may require adaptation for biofilms on other materials such as plastics or metals.
2. Biofilm formation: Ensure microbial cultures are fresh and in the exponential growth phase when preparing the inoculum for consistent biofilm formation. Variability in biofilm thickness may occur due to differences in species or growth media.
3. Slide handling: Handle glass slides gently during the rinsing and staining steps to avoid detachment of the biofilm (see Figure 4A). Avoid using force while dipping slides in water.
4. Stain drying: Do not air dry the slides for an extended period after applying stains, as this can lead to cracks forming due to over-drying (see Figure 4B).
5. Microscopy settings: Use a well-calibrated light microscope with 100× oil immersion for optimal visualization. Adjust the light source intensity and focus for clear differentiation of stained biofilm components.
6. Polymicrobial cultures: When preparing polymicrobial biofilms, ensure the initial inoculum is evenly mixed to avoid uneven colonization of the slide.
7. Contamination prevention: To avoid contamination of biofilms, especially by Bacillus species, avoid opening the Petri plate multiple times to add media (see Figure 4C). It is recommended to add enough media at the start to prevent the need for additional media during the incubation process.
8. Microscopy focus: During visualization of biofilms under the microscope, focusing especially on the corners of the biofilm can reveal better structural details, such as the honeycomb structure, which may be less apparent in the central regions.
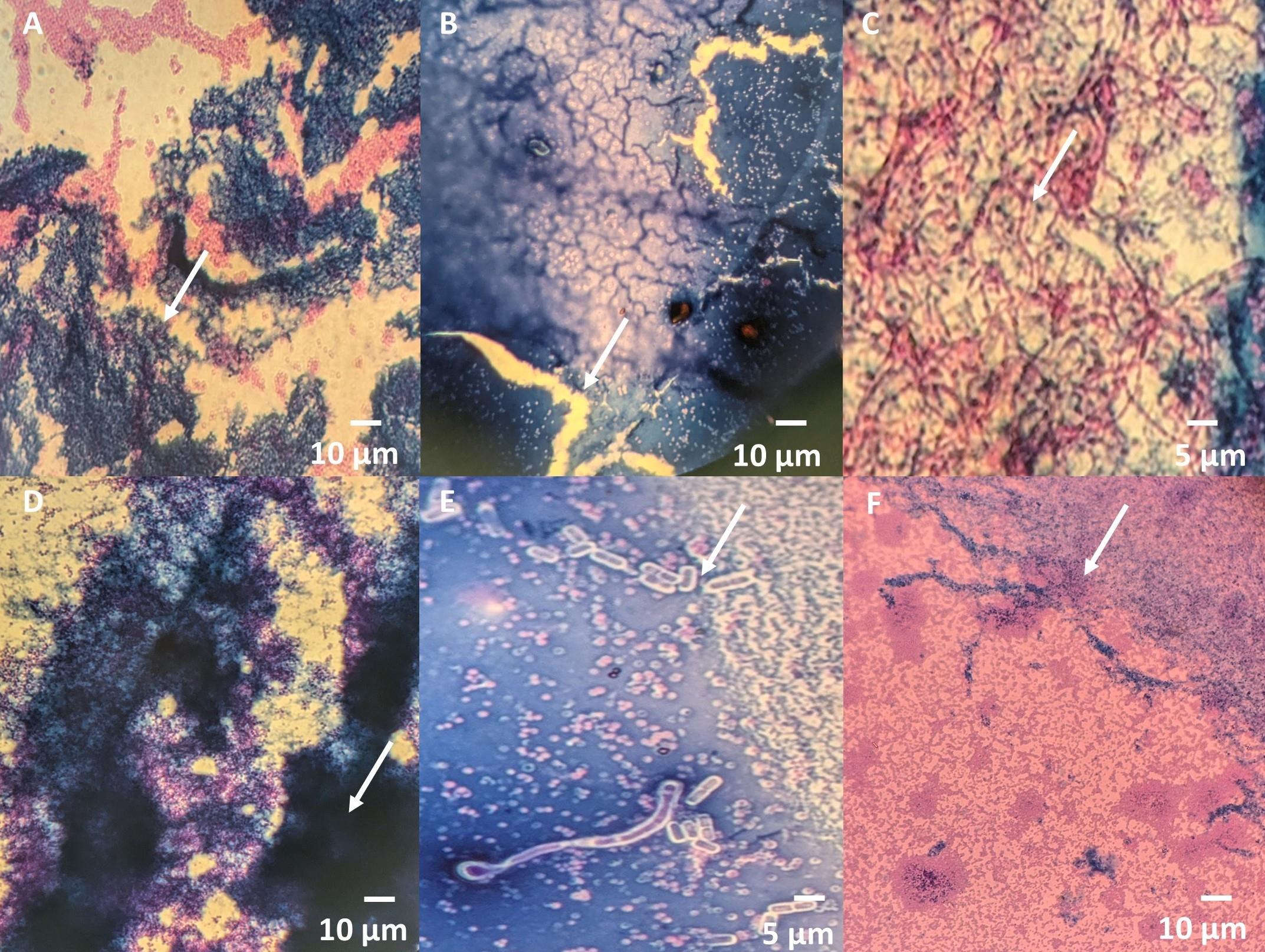
Figure 4. Common troubleshooting issues in biofilm detection using the dual-staining method. (A) Biofilm detachment. (B) Crack formation. (C) Contamination with Bacillus species. (D) Thick biofilm formation. (E) Dull or faded Maneval's staining of bacteria/yeast. (F) Improper Congo red staining of the polysaccharide matrix.
Troubleshooting (Table 1)
Table 1. Common troubleshooting issues encountered in the dual-staining method of biofilms using Maneval’s stain
| Problem | Possible Cause | Solution |
|---|---|---|
| 1. Biofilm detachment during rinsing (Figure 4A) | Excessive force during rinsing. | Gently dip the slide for 5 s in water without agitation. |
| 2. Weak or uneven staining | Inconsistent drying or inadequate stain incubation. | Ensure slides are completely dry but not over-dried before adding the next stain and adhere to incubation times. |
| 3. Cracks in stained biofilm (Figure 4B) | Over-drying of slides after staining. | Limit air-drying time and proceed to the next step after the slide is dry. |
| 4. Difficulty in biofilm visualization | Incorrect focus of microscope or insufficient stain adherence. | Ensure oil immersion is applied correctly and readjust focus. |
| 5. Contamination (Figure 4C) | Improper preparation or storage of cultures. | Use fresh cultures and maintain sterility during preparation. |
| 6. Variability in biofilm formation | Environmental factors or uneven inoculum. | Maintain consistent incubation conditions (temperature, humidity), and ensure the inoculum is well-mixed. |
| 7. Thick biofilm, unable to differentiate between bacteria and surrounding biofilm (see Figure 4D) | Too much inoculum taken during preparation. | Reduce the inoculum volume to achieve a thinner, clearer biofilm. |
| 8. Bacteria dull/faded appearance (see Figure 4E) | Insufficient incubation time for Maneval’s stain. | Ensure the slide is incubated with Maneval's stain for 10 min at room temperature for proper staining. |
| 9. Polysaccharide matrix of biofilm poorly stained (see Figure 4F) | Washing slides after Congo red staining. | Do not wash slides after applying Congo red; proceed directly to the next step. |
Acknowledgments
The authors acknowledge the Department of Biotechnology, Ministry of Science & Technology, Government of India, for Nirmala. B’s scholarship (DBT-JRF) and AIIMS Rishikesh for providing infrastructural support. The graphical abstract was created using Canva (Canva Pty Ltd, New South Wales, Australia). We also acknowledge Professor Richard P. Novick, Professor of Microbiology and Medicine at New York University, for providing the RN6390 strain. The authors gratefully acknowledge Edmond Maneval for the original discovery of Maneval’s stain in 1941 for staining bacterial capsules [11]. This protocol is based on an original article published in Scientific Reports (DOI: 10.1038/s41598-024-80644-3) [17].
Competing interests
The authors declare no conflicts of interest.
Ethical considerations
The study was approved by the Institutional Ethics Committee of All India Institute of Medical Sciences, Rishikesh (Approval Letter No: AIIMS/IEC/23/294).
References
- Ruijgrok, G., Wu, D. Y., Overkleeft, H. S. and Codée, J. D. C. (2024). Synthesis and application of bacterial exopolysaccharides. Curr Opin Chem Biol. 78. https://doi.org/10.1016/j.cbpa.2023.102418
- McLean, R. J. C., Lam, J. S. and Graham, L. L. (2012). Training the biofilm generation - A Tribute to J. W. Costerton. J Bacteriol. 194(24): 6706–6711. https://doi.org/10.1128/jb.01252-12
- Jamal, M., Ahmad, W., Andleeb, S., Jalil, F., Imran, M., Nawaz, M. A., et al. (2018). Bacterial biofilm and associated infections. J Chin Med Assoc. 81: 7–11. https://doi.org/10.1016/j.jcma.2017.07.012
- Algburi, A., Comito, N., Kashtanov, D. and Dicks, L. M. T., Chikindas, M. L. (2017). Control of biofilm formation: Antibiotics and beyond. Appl Environ Microbiol. 83. https://doi.org/10.1128/AEM.02508-16
- Abebe, G. M. (2020). The role of bacterial biofilm in antibiotic resistance and food contamination. Int J Microbiol. 2020. https://doi.org/10.1155/2020/1705814
- Di Pippo, F., Di Gregorio, L., Congestri, R., Tandoi, V. and Rossetti, S. (2018). Biofilm growth and control in cooling water industrial systems. FEMS Microbiol Ecol. 94. https://doi.org/10.1093/femsec/fiy044
- Romero, F., Acuña, V. and Sabater, S. (2020). Multiple stressors determine community structure and estimated function of river biofilm bacteria. Appl Environ Microbiol. https://doi.org/10.1128/AEM
- Mohammed, Y. H. E., Manukumar, H. M., Rakesh, K. P., Karthik, C. S., Mallu, P. and Qin, H. L. (2018). Vision for medicine: Staphylococcus aureus biofilm war and unlocking keys for anti-biofilm drug development. Microb Pathog. 123: 339–347. https://doi.org/10.1016/j.micpath.2018.07.002
- Wilson, C., Lukowicz, R., Merchant, S., Valquier-Flynn, H., Caballero, J., Sandoval, J., et al. (2017). Quantitative and qualitative assessment methods for biofilm growth: A mini-review. Microbiol Spectrum. 6. https://pmc.ncbi.nlm.nih.gov/articles/PMC6133255/
- Achinas, S., Yska, S. K., Charalampogiannis, N., Krooneman, J. and Euverink, G. J. W. (2020). A technological understanding of biofilm detection techniques: A review. Materials. 13. https://doi.org/10.3390/ma13143147
- Dunstan, R. A., Bamert, R. S., Belousoff, M. J., Short, F. L., Barlow, C. K., Pickard, D. J., et al. (2021). Mechanistic insights into the capsule-targeting depolymerase from a Klebsiella pneumoniae bacteriophage. Microbiol Spectrum. 9(1). https://doi.org/10.1128/spectrum.01023-21
- Yuguchi, Y., Hirotsu, T. and Hosokawa, J. (2005). Structural characteristics of xyloglucan - Congo red aggregates as observed by small angle X-ray scattering. Cellulose. 12(5): 469–477. https://doi.org/10.1007/s10570-005-4434-7
- Kashyout, A. H., Soliman, H., Nabil, M. and Bishara, A. (2015). Impact of Congo red dye in nano-porous silicon as pH-sensor. Sens Actuators B Chem. 216: 279–285. https://doi.org/10.1016/j.snb.2015.03.099
- Hughes, R. B. and Smith, A. C. (2007). Capsule stain protocols. Am Soc Microbiol.
- Pelzer, E. S., Allan, J. A., Theodoropoulos, C., Ross, T., Beagley, K. W. and Knox, C. L. (2012). Hormone-dependent bacterial growth, persistence, and biofilm formation–a pilot study investigating human follicular fluid collected during IVF cycles. PLOS One. 7(12): e49965. https://doi.org/10.1371/journal.pone.0049965
- Ito A, Taniuchi A, May T, Kawata K, Okabe S. (2009). Increased antibiotic resistance of Escherichia coli in mature biofilms. Appl Environ Microbiol. 75(12): 4093–4100. https://doi.org/10.1128/AEM.02949-08
- B, N., Manhas, P. L., Jadli, M., Sharma, R., Manhas, H. and Omar, B. J. (2024). A novel dual-staining method for cost-effective visualization and differentiation of microbial biofilms. Sci Rep. 14(1): 29169. https://doi.org/10.1038/s41598-024-80644-3
Article Information
Publication history
Received: Dec 12, 2024
Accepted: Jan 31, 2025
Available online: Feb 24, 2025
Published: Mar 5, 2025
Copyright
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Nirmala, B. and Omar, B. J. (2025). Microbial Biofilm Detection and Differentiation by Dual Staining Using Maneval’s Stain. Bio-protocol 15(5): e5228. DOI: 10.21769/BioProtoc.5228.
- B, N., Manhas, P. L., Jadli, M., Sharma, R., Manhas, H. and Omar, B. J. (2024). A novel dual-staining method for cost-effective visualization and differentiation of microbial biofilms. Sci Rep. 14(1): 29169. https://doi.org/10.1038/s41598-024-80644-3
Category
Microbiology > Microbial biofilm > Biofilm detection
Microbiology > Microbial cell biology > Cell staining
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Tips for asking effective questions
+ Description
Write a detailed description. Include all information that will help others answer your question including experimental processes, conditions, and relevant images.
Share
Bluesky
X
Copy link








