- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
An Activity-Based Proteomics with Two-Dimensional Polyacrylamide Gel Electrophoresis (2D-PAGE) for Identifying Target Proteases in Arabidopsis Apoplastic Fluid
Published: Vol 15, Iss 5, Mar 5, 2025 DOI: 10.21769/BioProtoc.5226 Views: 1949
Reviewed by: Min CaoAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
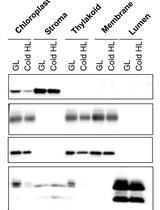
A Simple Sonication Method to Isolate the Chloroplast Lumen in Arabidopsis thaliana
Jingfang Hao and Alizée Malnoë
Aug 5, 2023 2263 Views
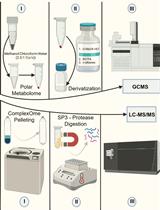
Streamlining Protein Fractional Synthesis Rates Using SP3 Beads and Stable Isotope Mass Spectrometry: A Case Study on the Plant Ribosome
Dione Gentry-Torfer [...] Federico Martinez-Seidel
May 5, 2024 2843 Views
Advancing 2-DE Techniques: High-Efficiency Protein Extraction From Lupine Roots
Sebastian Burchardt [...] Emilia Wilmowicz
Oct 5, 2025 1736 Views
Abstract
Plant proteases participate in a wide variety of biological processes, including development, growth, and defense. To date, numerous proteases have been functionally identified through genetic studies. However, redundancy among certain proteases can obscure their roles, as single-gene loss-of-function mutants often exhibit no discernible phenotype, limiting identification through genetic approaches. Here, we describe an efficient system for the identification of target proteases that cleave specific substrates in the Arabidopsis apoplastic fluid. The method involves using Arabidopsis-submerged culture medium, which contains apoplastic proteases, followed by native two-dimensional electrophoresis. Gel fractionation and an in-gel peptide cleavage assay with a fluorescence-quenching peptide substrate are then used to detect specific proteolytic activity. The active fraction is then subjected to mass spectrometry–based proteomics to identify the protease of interest. This method allows for the efficient and comprehensive identification of proteases with specific substrate cleavage activities in the apoplast.
Key features
• Targets Arabidopsis thaliana secreted protease but may be applicable to other plant species and intracellular proteases if protease-enriched samples are available.
• The protocol involves an in-gel peptide cleavage assay of native two-dimensional gels diced with SAINOME plates, using a fluorescence-quenching substrate.
• Facilitates the efficient identification of proteases with the desired activity from the entire sample, without restricting the analysis to a specific class of proteases.
Keywords: Extracellular proteaseGraphical overview
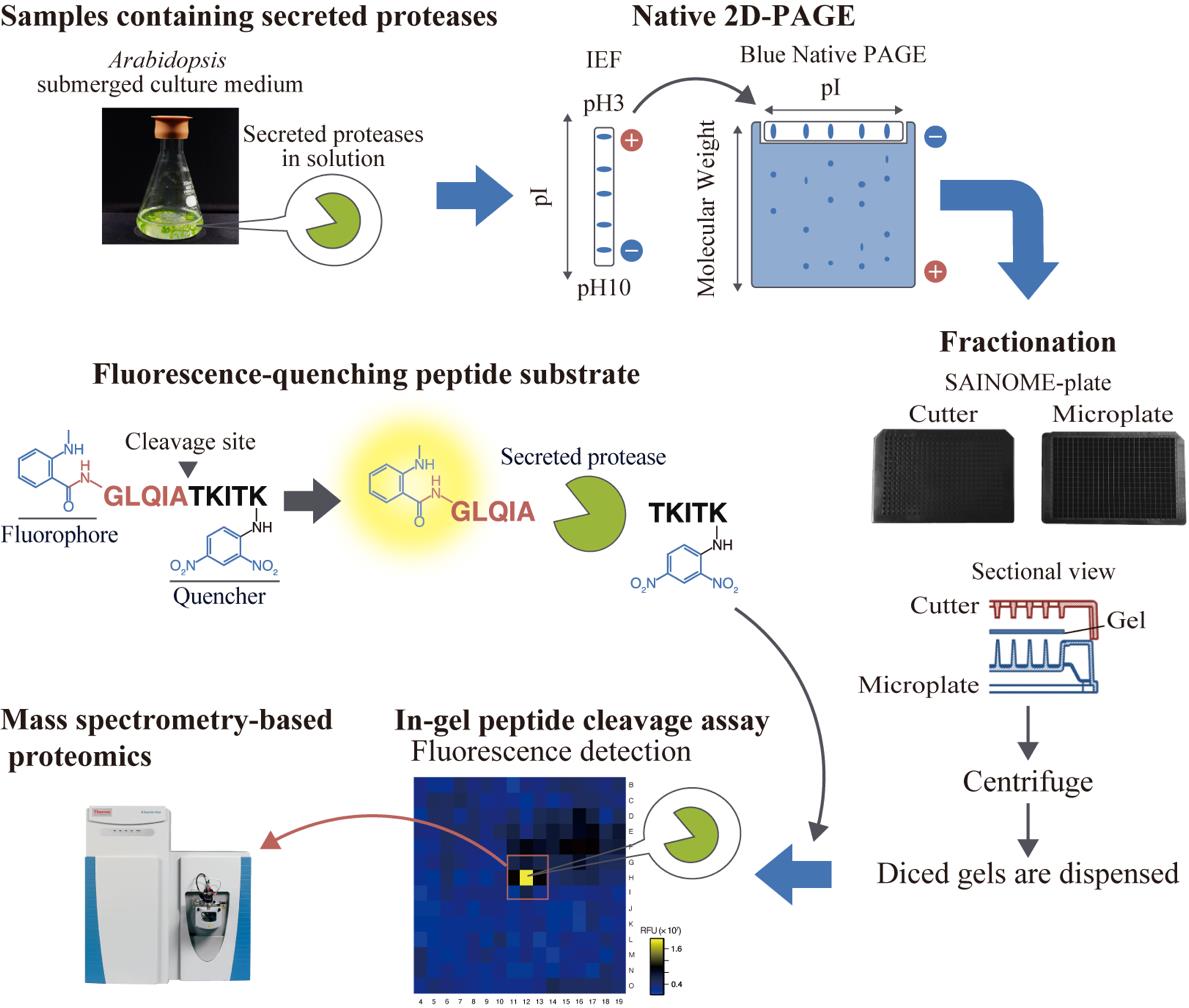
High-throughput system for identifying proteases that cleave specific substrates in Arabidopsis apoplastic fluid. Arabidopsis-submerged culture medium containing secreted proteases is subjected to native two-dimensional electrophoresis. After electrophoresis, the gel is fractionated using a SAINOME plate, and an in-gel peptide cleavage assay is performed using a fluorescence-quenching peptide substrate that fluorescently detects proteolytic activity at the specific cleavage site. Mass spectrometry–based proteomics is performed to identify proteases in the active fraction.
Background
Plant proteases participate in a variety of physiological processes, including development, growth, and defense [1]. Their roles extend beyond protein degradation to include protein activation and regulation of protein localization through limited and site-specific proteolysis [2]. Among the numerous proteases, secreted proteases in the apoplast have received increasing attention in recent years, mainly because of their roles in immunity and peptide hormone processing [3–5]. In the model plant Arabidopsis thaliana, more than 800 putative protease genes have been identified, nearly 300 of which encode secreted proteases with signal peptides [1,6,7].
Proteases often exhibit functional redundancy, which complicates their study through genetic approaches. To address this issue, some studies have employed inhibitors that specifically target particular protease families and suppress their function [8–11]. Alternatively, strategies focusing on changes in transcript levels or enzyme activity under specific conditions have been used to identify proteases of interest [12,13]. Protease identification has also been attempted by narrowing down candidate proteases based on the similarity between the cleavage site of a target substrate and the cleavage sequence of a known protease [14,15]. However, these approaches are all indirect and do not directly assess the specificity of the recognition sequence of the enzyme. Consequently, these methods are not always effective for identifying the protease responsible for cleaving a specific substrate protein.
We have developed a high-throughput system for identifying proteases involved in the cleavage of specific substrates at defined cleavage sites from samples containing apoplastic proteases. This approach employs native two-dimensional (2D) electrophoresis, followed by gel dicing using a SAINOME plate and an in-gel peptide cleavage assay with a fluorescence-quenching peptide substrate [16–19]. Using this system, we recently identified secreted proteases involved in the C-terminal cleavage of the immunogenic peptide flg22, derived from the bacterial flagellar protein flagellin [20]. Here, we provide a step-by-step method for identifying secreted proteases that cleave specific substrates at defined sites, using the identification of flagellin-cleaving proteases as an example. This method is broadly applicable to any proteases that retain enzymatic activity following native 2D electrophoresis.
Materials and reagents
Biological materials
1. Arabidopsis thaliana accession Col-0 submerged culture medium [20] (see General note 1)
Reagents
1. Gamborg's B5 medium salt mixture (Shiotani M.S., catalog number: 399-00621)
2. Sucrose (Wako, catalog number: 199-00027)
3. Nicotinic acid (Nacalai Tesque, catalog number: 24326-52)
4. Pyridoxine hydrochloride (Wako, catalog number: 165-05401)
5. Thiamin hydrochloride (Wako, catalog number: 201-00852)
6. Myo-inositol (Sigma-Aldrich, catalog number: I5125)
7. Glycine (Wako, catalog number: 077-00735)
8. Agar (Wako, catalog number: 016-11875)
9. Spectra/Por 7 membrane (molecular weight cutoff 10 kDa) (Repligen, catalog number: 132117)
10. 3-[(3-Cholamidopropyl)dimethylammonio]propanesulfonate (CHAPS) (Dojindo, catalog number: C008)
11. Immobilized pH gradient (IPG) buffer, pH 3–10 NL (GE Healthcare, catalog number: 17-6000-88)
12. (+/-)-Dithiothreitol (DTT) (Wako, catalog number: 049-08972)
13. Bromophenol blue (BPB) (Wako, catalog number: 021-02911)
14. Immobiline DryStrip pH 3–10 NL, 7 cm (IPG strip) (Cytiva, catalog number: 17-6001-12)
15. Immobiline DryStrip cover fluid (Cytiva, catalog number: 17-1335-01)
16. 6-Aminocaproic acid (LKT Laboratories, catalog number: A4935)
17. Glycerol (Nacalai Tesque, catalog number: 17018-25)
18. 2-Amino-2-hydroxymethyl-1,3-propanediol (Tris) (Wako, catalog number: 204-07885)
19. Coomassie brilliant blue G 250 (CBB G-250) (Fluka, catalog number: 27815)
20. NativePAGE running buffer (20×) (Invitrogen, catalog number: BN2001)
21. NativePAGE 4%–16%, Bis-Tris, 1.0 mm, mini protein gels, 15-well (Invitrogen, catalog number: BN1004BOX)
22. NativeMarkTM unstained protein standard (Invitrogen, catalog number: LC0725)
23. Nma-flg[47–56]-Dnp [fluorescence-quenching substrate, Nma-GLQIATKITK(Dnp)-NH2] (Peptide Institute, custom-made) (see General Note 2)
24. 2-Morpholinoethanesulfonic acid, monohydrate (MES) (Dojindo, catalog number: GB12)
25. Methanol (Wako, catalog number: 131-01826)
26. Acetic acid (Wako, catalog number: 017-00256)
27. Lysyl endopeptidase (Lys-C) (Wako, catalog number: 125-05061)
28. Trypsin (Promega, catalog number: V5111)
Solutions
1. B5 vitamin solution (100×) (see Recipes)
2. B5 medium (see Recipes)
3. Rehydration solution A (see Recipes)
4. Rehydration solution B (see Recipes)
5. Rehydration solution (see Recipes)
6. Equilibration solution (see Recipes)
7. Native PAGE anode buffer (see Recipes)
8. Native PAGE cathode buffer (see Recipes)
9. In-gel peptide cleavage assay solution (see Recipes)
10. Fixing solution (see Recipes)
Recipes
1. B5 vitamin solution (100×)
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Nicotinic acid | 50 mg/L | 25 mg |
| Pyridoxine hydrochloride | 50 mg/L | 25 mg |
| Thiamin hydrochloride | 10 mg/L | 5 mg |
| Myo-inositol | 10 g/L | 5 g |
| Glycine | 200 mg/L | 100 mg |
| Milli-Q water | n/a | Up to 500 mL |
| Total | n/a | 500 mL |
2. B5 medium
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Gamborg's B5 medium salt mixture | n/a | One bag (3.3 g) |
| B5 vitamin solution (100×) | 1× | 10 mL |
| Sucrose | 1% (w/v) | 10 g |
| Milli-Q water | n/a | Up to 1 L |
| Total | n/a | 1 L |
Adjust pH to 5.7 with 1 N KOH. Autoclave at 121 °C for 20 min. For solid medium, add 7 g of agar before autoclaving.
3. Rehydration solution A
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| CHAPS | 4% (w/v) | 40 mg |
| IPG buffer | 0.5% (v/v) | 5 μL |
| DTT (15.9 mg/mL) | 0.62 mg/mL | 39 μL |
| Milli-Q water | n/a | Up to 1 mL |
| Total | n/a | 1 mL |
4. Rehydration solution B
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| CHAPS | 2% (w/v) | 20 mg |
| IPG buffer | 0.5% (v/v) | 5 μL |
| BPB [0.05% (w/v)] | 0.002% (w/v) | 40 μL |
| Milli-Q water | n/a | Up to 1 mL |
| Total | n/a | 1 mL |
5. Rehydration solution
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Rehydration solution A | 12.5% (v/v) | 20 μL |
| Rehydration solution B | 87.5% (v/v) | 140 μL |
| Total | n/a | 160 μL |
6. Equilibration solution
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| 6-Aminocaproic acid | 450 mM | 0.59 g |
| Glycerol | 25.5% (v/v) | 2.55 mL |
| Tris-HCl (0.5 M, pH 6.8) | 50 mM | 1 mL |
| CBB G-250 | 0.02% (w/v) | 2 mg |
| Milli-Q water | n/a | Up to 10 mL |
| Total | n/a | 10 mL |
7. Native PAGE anode buffer
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| 20× Native PAGE running buffer | 1× | 10 mL |
| Milli-Q water | n/a | Up to 200 mL |
| Total | n/a | 200 mL |
8. Native PAGE cathode buffer
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| 20× Native PAGE running buffer | 1× | 5 mL |
| CBB G-250 [0.4% (w/v)] | 0.002% (w/v) | 500 μL |
| Milli-Q water | n/a | Up to 100 mL |
| Total | n/a | 100 mL |
9. In-gel peptide cleavage assay solution
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Nma-flg[47–56]-Dnp (0.4 mM) | 40 μM | 1.7 mL |
| MES-KOH (pH5.7) | 25 mM | 850 μL |
| Milli-Q water | n/a | 14.45 mL |
| Total | n/a | 17 mL |
10. Fixing solution
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Methanol | 20% (v/v) | 20 mL |
| Acetic acid | 7.5% (v/v) | 7.5 mL |
| Milli-Q water | n/a | 72.5 mL |
| Total | n/a | 100 mL |
Laboratory supplies
1. 300 mL flask (Iwaki, catalog number: 4980FK300)
2. 15 mL centrifuge tubes (Corning, catalog number: 430791)
3. 1.5 mL microtubes (e.g., BMBio, catalog number: BM-15F)
4. Stainless steel surgical blade, No. 14 (FEATHER Safety Razor, model: No. 14)
5. Tweezers (e.g., rubisTech, catalog number: SS-SA)
6. Square Petri dish (Eiken Chemical, catalog number: AW2000)
7. SAINOME, black [SAINOME consists of a microplate (384-well) and a cutter] (Sainome, catalog number: SAI-386-B)
Equipment
1. Rotary vacuum evaporator (EYELA, model: N-N series)
2. High-speed micro centrifuge CF15RN (Hitachi Koki, model: himac CF15RN)
3. Angle rotor (Hitachi Koki, model: T11A31)
4. Angle rotor (Hitachi Koki, model: T15AP31)
5. Strip holder, 7 cm (Cytiva, catalog number: 80-6416-87)
6. Ettan IPGphor II isoelectric focusing unit (Amersham Biosciences, catalog number: 80-6505-03)
7. Mini-slab electrophoretic apparatus (Bio Craft, model: BE-211)
8. High-speed micro centrifuge CF16RN (Hitachi Koki, model: himac CF16RN)
9. Swing rotor (Koki Holdings, model: T5S32)
10. VICTOR Nivo multimode plate reader equipped with a 340/20 nm excitation filter and a 435/20 nm emission filter (PerkinElmer, catalog number: HH35000500)
11. 340/20 nm filter (PerkinElmer, catalog number: HH35000912)
12. 435/20 nm filter (PerkinElmer, catalog number: HH35000918)
13. Extrahera (Biotage, catalog number: 414001)
14. Mass spectrometer (Thermo Fisher Scientific, model: Q Exactive Hybrid Quadrupole-Orbitrap Mass Spectrometer)
15. Gradient pump (Thermo Fisher Scientific, model: Dionex U3000)
Software and datasets
1. VICTOR Nivo software ver. 4.0 (PerkinElmer, 2020 May)
2. Proteome Discoverer ver. 2.3 (Thermo Fisher Scientific)
Procedure
A. Native 2D electrophoresis
1. Sample preparation
a. Prepare Arabidopsis whole-plant-submerged culture as described previously [20]. Briefly, transplant forty 6-day-old seedlings, germinated on B5 solid medium, into 100 mL of B5 liquid medium in a 300 mL flask. Incubate them for 7–11 days at 22 °C under continuous light without shaking.
Note: The flask should not be continuously shaken, but it is recommended to gently shake it by hand a few times a day.
b. Collect the spent medium by decantation (Arabidopsis-submerged culture medium).
c. Concentrate 100 mL of the Arabidopsis-submerged culture medium to 10 mL using a rotary evaporator with the water bath set to 37 °C.
d. Transfer the concentrated sample into 15 mL centrifuge tubes and centrifuge at 11,900× g for 5 min using a himac CF15RN (T11A31 rotor).
e. Dialyze the supernatant overnight with stirring at 4 °C against 3 L of reverse osmosis (RO) water using a Spectra/Por 7 membrane.
f. Transfer the dialyzed sample to 15 mL centrifuge tubes and centrifuge at 11,900× g for 5 min using a himac CF15RN (T11A31 rotor).
g. Transfer the supernatant to a new 15 mL centrifuge tube, freeze at -80 °C, and lyophilize.
h. Dissolve the lyophilized powder in 125 μL of rehydration solution and transfer to a 1.5 mL tube.
2. Native isoelectric focusing (IEF)
a. Centrifuge 125 μL of the dehydration solution containing the sample at 18,800× g for 5 min using a himac CF15RN (T15AP31 rotor).
b. Load the supernatant into a 7 cm strip holder.
c. Place the immobilized pH gradient (IPG strip) in the strip holder with the gel-side down in the dehydration solution.
d. Apply DryStrip cover fluid to the strip and cover the strip holder with the plastic lid.
e. Place the strip holder on the Ettan IPGphor platform and perform IEF using the following program:
Rehydrate for 14 h at 20 °C.
Perform IEF at 50 μA/strip and 20 °C using the following steps:
Step 1: 500 V for 30 min
Step 2: 1,000 V for 30 min
Step 3: 5,000 V for 100 min
Note: The maximum voltage may not be reached during the program. The rehydration time can be adjusted for convenience but must be longer than 10 h.
3. Blue native polyacrylamide gel electrophoresis (PAGE)
a. After IEF, place the IPG strip in 10 mL of equilibration solution in a 15 mL tube.
b. Equilibrate the IPG strip at 4 °C for 15 min.
c. Rinse the IPG strip with native PAGE anode buffer.
d. Remove wells from the native PAGE gel using a surgical blade and tweezers to prepare a gel with one protein marker well and one wide well.
e. Using tweezers, place the IPG strip in the wide well and gently press the strip onto the top surface of the native PAGE gel (Figure 1).
Note: Slightly trim the ends of the strip to fit into the well. Avoid air bubbles between the strip and the gel.
f. Place the gel into the electrophoretic apparatus and pour native PAGE cathode buffer into the upper buffer tank and native PAGE anode buffer into the lower buffer tank.
g. Load 3 μL of NativeMark unstained protein standard into the protein marker well.
h. Run electrophoresis at 150 V for approximately 90 min or until the dye front line reaches the end of the gel.

Figure 1. Preparation for native PAGE. Native PAGE gel with an IPG strip placed on top.
B. In-gel peptide cleavage assay
1. Fractionation of the gel after electrophoresis using a SAINOME plate
a. Remove the top and bottom of the gel with a surgical blade, taking care to remove the contact surface with the IPG strip and the area below the dye front. Also, remove the marker lane (Figure 2A).
Note: The marker lane can be used as a reference for evaluating native PAGE separation and may be stained or destained as appropriate.
b. Rinse the gel with 25 mM MES-KOH pH 5.7 in a square Petri dish.
c. Place the gel on a SAINOME 384-well microplate (Figure 2B).
d. Position the SAINOME cutter, which is designed to fit on top of the plate, on the gel.
e. Press the cutter firmly onto the gel and secure the plate and cutter together with tape (Figure 2C).
f. Centrifuge the assembly at 2,000× g for 5 min using a himac CF16RN (T5S32 rotor).
Note: By centrifugation, the gel is cut into 4.5 mm squares and dispensed into the 384-well plate.

Figure 2. Fractionation of the gel using the SAINOME plate. A. Gel after native two-dimensional electrophoresis. Remove all parts of the gel except the part enclosed by the red square. B. Placement of the gel on the SAINOME plate. C. SAINOME plate with cutter positioned and secured with tape.
2. In-gel peptide cleavage assay of fluorescence-quenching substrate
a. Remove the cutter from the plate and add 60 μL of in-gel peptide cleavage assay solution to each well containing gel pieces.
b. Measure the fluorescence intensity in each well using a plate reader with an excitation wavelength of 340 nm and an emission wavelength of 435 nm every hour for 16 h (Figure 3).
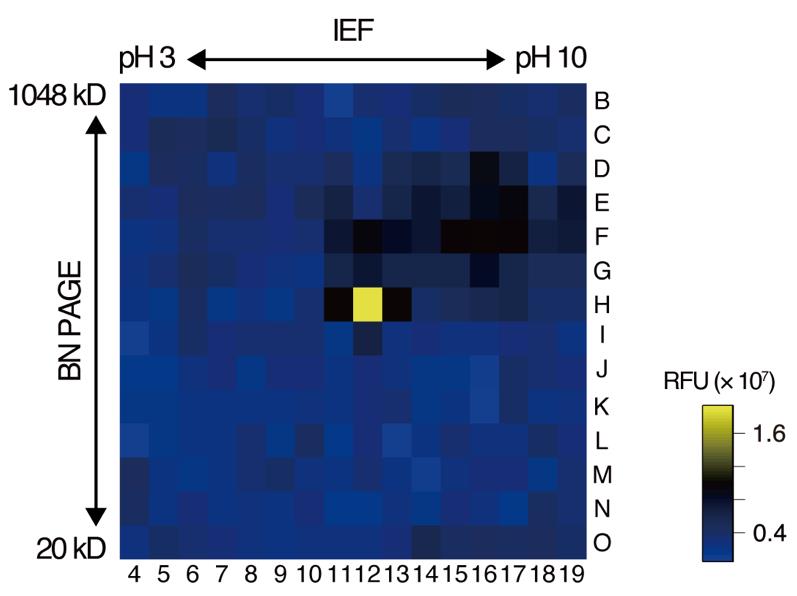
Figure 3. Representative results of the in-gel peptide cleavage assay using a fluorescence-quenching substrate. Fluorescence intensity 16 h after substrate addition is shown as an example. The numbers and letters represent the columns and rows, respectively, of the microplate used in the assay. Fluorescence intensity is color-coded according to the scale shown on the right. BN PAGE: blue native polyacrylamide gel electrophoresis; IEF: isoelectric focusing; RFU: relative fluorescence units.
C. Proteomic analysis of the active gel fractions
1. In-gel protein digestion
a. Collect assay solution and gel fragments separately from the fluorescence-positive and control fluorescence-negative wells and place each into separate 1.5 mL tubes.
Note: Nano-liquid chromatography–mass spectrometry (nano-LC–MS) analysis of the collected assay solution can confirm whether correct cleavage of the substrate occurred.
b. Fix the gel fragments with fixing solution at room temperature for 1 h.
c. Transfer the fixed gel fragments to a 96-well filter plate with tweezers and perform reduction and alkylation using Extrahera, as described previously [21].
d. Transfer the gel fragments with tweezers to a new 1.5 mL tube and digest with Lys-C/trypsin as previously described [21].
2. Nano-liquid chromatography–tandem mass spectrometry (nano-LC–MS/MS) analysis
a. Analyze 7.25 μL aliquots by nano-LC–MS/MS using a system combining a Q Exactive Hybrid Quadrupole-Orbitrap mass spectrometer with a Dionex U3000 gradient pump as described previously [21].
3. MS/MS data analysis
a. Process and analyze the MS/MS raw files with Proteome Discoverer 2.3 using the SEQUEST HT algorithm, searching against the TAIR10 Arabidopsis protein database.
b. Use the following search parameters: peptide mass range, 350–5,000 Da; enzyme specificity, trypsin (Full), with up to two missed cleavages; minimum peptide length, six residues; maximum equal modifications per peptide, 3; precursor ion and peptide fragment mass tolerances, ±10 ppm and ±0.02 Da, respectively; static modification, carbamidomethyl (Cys); and dynamic modifications, oxidation (Met).
c. Extract proteases with signal peptides from the resulting high-confidence master protein list using TAIR Gene Ontology (GO) and SignalP 6.0 [7].
d. Identify secreted proteases specifically present in the active fraction based on peptide-spectrum match (PSM) and abundance scores.
Validation of protocol
This protocol or parts of it has been used and validated in the following research article:
• Matsui et al. [20]. Arabidopsis SBT5.2 and SBT1.7 subtilases mediate C-terminal cleavage of flg22 epitope from bacterial flagellin. Nature Communications (Figure 3, panels a and b).
General notes and troubleshooting
General notes
1. Biological material: Although Arabidopsis-submerged culture medium is used as an example, this method is not limited to Arabidopsis samples. Any sample in which proteases retain activity after native 2D electrophoresis can be used.
2. Fluorescence-quenching peptide substrate: Nma-flgN[47–56]-Dnp, with a purity of 97.4%, specifically detects proteolytic activity at the C-terminus of the flg22 domain [20]. A paired fluorophore and quencher are introduced at opposite ends of the peptide substrate. Hydrolysis of an internal peptide bond increases the distance between the donor and acceptor, preventing quenching of the fluorophore by the quencher and enhancing the fluorescence signal. The proteolytic activity of other proteases can also be detected by altering the amino acid sequence of the peptide substrate.
Acknowledgments
This protocol was originally described and validated in Matsui et al. [20].
This research was supported by a Grant-in-Aid for Scientific Research (S) (No. 23H05477 to Y.M.), a Grant-in-Aid for Transformative Research Areas (A) (No. 20H05907 to Y.M.), and the Nagoya University Interdisciplinary Frontier Fellowship (to S.M.) supported by Nagoya University and the Japan Science and Technology Agency (JST) (No. JPMJFS2120).
Competing interests
The authors declare no conflict of interest.
References
- van der Hoorn, R. A. (2008). Plant proteases: from phenotypes to molecular mechanisms. Annu Rev Plant Biol. 59: 191–223. https://doi.org/10.1146/annurev.arplant.59.032607.092835
- Schaller, A. (2004). A cut above the rest: the regulatory function of plant proteases. Planta. 220(2): 183–197. https://doi.org/10.1007/s00425-004-1407-2
- Godson, A. and van der Hoorn, R. A. L. (2021). The front line of defence: a meta-analysis of apoplastic proteases in plant immunity. J Exp Bot. 72(9): 3381–3394. https://doi.org/10.1093/jxb/eraa602
- Stührwohldt, N. and Schaller, A. (2019). Regulation of plant peptide hormones and growth factors by post-translational modification. Plant Biol. 21: 49–63. https://doi.org/10.1111/plb.12881
- Stintzi, A. and Schaller, A. (2022). Biogenesis of post-translationally modified peptide signals for plant reproductive development. Curr Opin Plant Biol. 69: 102274. https://doi.org/10.1016/j.pbi.2022.102274
- Rawlings, N. D., Barrett, A. J., Thomas, P. D., Huang, X., Bateman, A. and Finn, R. D. (2018). The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Res. 46(D1): D624–D632. https://doi.org/10.1093/nar/gkx1134
- Teufel, F., Almagro Armenteros, J. J., Johansen, A. R., Gislason, M. H., Pihl, S. I., Tsirigos, K. D., Winther, O., Brunak, S., von Heijne, G. and Nielsen, H. (2022). SignalP 6.0 predicts all five types of signal peptides using protein language models. Nat Biotechnol. 40(7): 1023–1025. https://doi.org/10.1038/s41587-021-01156-3
- Schardon, K., Hohl, M., Graff, L., Pfannstiel, J., Schulze, W., Stintzi, A. and Schaller, A. (2016). Precursor processing for plant peptide hormone maturation by subtilisin-like serine proteinases. Science. 354(6319): 1594–1597. https://doi.org/10.1126/science.aai8550
- Stuhrwohldt, N., Scholl, S., Lang, L., Katzenberger, J., Schumacher, K. and Schaller, A. (2020). The biogenesis of CLEL peptides involves several processing events in consecutive compartments of the secretory pathway. eLife. 9: e55580. https://doi.org/10.7554/eLife.55580
- Paulus, J. K., Kourelis, J., Ramasubramanian, S., Homma, F., Godson, A., Horger, A. C., Hong, T. N., Krahn, D., Ossorio Carballo, L., Wang, S., et al. (2020). Extracellular proteolytic cascade in tomato activates immune protease Rcr3. Proc Natl Acad Sci USA. 117(29): 17409–17417. https://doi.org/10.1073/pnas.1921101117
- Stuhrwohldt, N., Ehinger, A., Thellmann, K. and Schaller, A. (2020). Processing and formation of bioactive CLE40 peptide are controlled by posttranslational proline hydroxylation. Plant Physiol. 184(3): 1573–1584. https://doi.org/10.1104/pp.20.00528
- Wang, Y., Garrido-Oter, R., Wu, J., Winkelmuller, T. M., Agler, M., Colby, T., Nobori, T., Kemen, E. and Tsuda, K. (2019). Site-specific cleavage of bacterial MucD by secreted proteases mediates antibacterial resistance in Arabidopsis. Nat Commun. 10(1): 2853. https://doi.org/10.1038/s41467-019-10793-x
- Morimoto, K. and van der Hoorn, R. A. (2016). The increasing impact of activity-based protein profiling in plant science. Plant Cell Physiol. 57(3): 446–461. https://doi.org/10.1093/pcp/pcw003
- Wrzaczek, M., Vainonen, J. P., Stael, S., Tsiatsiani, L., Help-Rinta-Rahko, H., Gauthier, A., Kaufholdt, D., Bollhoner, B., Lamminmaki, A., Staes, A., et al. (2015). GRIM REAPER peptide binds to receptor kinase PRK5 to trigger cell death in Arabidopsis. EMBO J. 34(1): 55–66. https://doi.org/10.15252/embj.201488582
- Yang, H., Kim, X., Sklenar, J., Aubourg, S., Sancho-Andres, G., Stahl, E., Guillou, M. C., Gigli-Bisceglia, N., Tran Van Canh, L., Bender, K. W., et al. (2023). Subtilase-mediated biogenesis of the expanded family of SERINE RICH ENDOGENOUS PEPTIDES. Nat Plants. 9(12): 2085–2094. https://doi.org/10.1038/s41477-023-01583-x
- Komatsu, T., Hanaoka, K., Adibekian, A., Yoshioka, K., Terai, T., Ueno, T., Kawaguchi, M., Cravatt, B. F. and Nagano, T. (2013). Diced electrophoresis gel assay for screening enzymes with specified activities. J Am Chem Soc. 135(16): 6002–6005. https://doi.org/10.1021/ja401792d
- Komatsu, T., Shimoda, M., Kawamura, Y., Urano, Y. and Nagano, T. (2019). Development and validation of an improved diced electrophoresis gel assay cutter-plate system for enzymomics studies. Biochim Biophys Acta Proteins Proteom. 1867(1): 82–87. https://doi.org/10.1016/j.bbapap.2018.06.004
- Matayoshi, E. D., Wang, G. T., Krafft, G. A. and Erickson, J. (1990). Novel fluorogenic substrates for assaying retroviral proteases by resonance energy transfer. Science. 247(4945): 954–958. https://doi.org/10.1126/science.2106161
- Bickett, D. M., Green, M. D., Berman, J., Dezube, M., Howe, A. S., Brown, P. J., Roth, J. T. and McGeehan, G. M. (1993). A high throughput fluorogenic substrate for interstitial collagenase (MMP-1) and gelatinase (MMP-9). Anal Biochem. 212(1): 58–64. https://doi.org/10.1006/abio.1993.1291
- Matsui, S., Noda, S., Kuwata, K., Nomoto, M., Tada, Y., Shinohara, H. and Matsubayashi, Y. (2024). Arabidopsis SBT5.2 and SBT1.7 subtilases mediate C-terminal cleavage of flg22 epitope from bacterial flagellin. Nat Commun. 15(1): 3762. https://doi.org/10.1038/s41467-024-48108-4
- Kano, K., Noda, S., Sato, S., Kuwata, K. and Mishiro‐Sato, E. (2023). An efficient in‐gel digestion method on small amounts of protein sample from large intact gel pieces. Sep Sci Plus. 6(4). https://doi.org/10.1002/sscp.202200121
Article Information
Publication history
Received: Dec 9, 2024
Accepted: Jan 25, 2025
Available online: Feb 16, 2025
Published: Mar 5, 2025
Copyright
© 2025 The Author(s); This is an open access article under the CC BY license (https://creativecommons.org/licenses/by/4.0/).
How to cite
Matsui, S. and Matsubayashi, Y. (2025). An Activity-Based Proteomics with Two-Dimensional Polyacrylamide Gel Electrophoresis (2D-PAGE) for Identifying Target Proteases in Arabidopsis Apoplastic Fluid. Bio-protocol 15(5): e5226. DOI: 10.21769/BioProtoc.5226.
Category
Plant Science > Plant biochemistry > Protein > Isolation and purification
Biochemistry > Protein > Electrophoresis
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










