- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Flood Inoculation of Fusarium eumartii in Tomato Seedlings: Method for Evaluating the Infectivity of Pathogen Spores
Published: Vol 15, Iss 3, Feb 5, 2025 DOI: 10.21769/BioProtoc.5198 Views: 1586
Reviewed by: Noelia ForesiMalgorzata Lichocka

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

A Reliable In Planta Inoculation and Antifungal Screening Protocol for Rhizoctonia solani-Induced Sheath Blight in Rice
Alinaj Yasin [...] Palash Deb Nath
Nov 5, 2025 1584 Views

In Silico Prediction and In Vitro Validation of Bacterial Interactions in the Plant Rhizosphere Using a Synthetic Bacterial Community
Arijit Mukherjee [...] Sanjay Swarup
Nov 5, 2025 1671 Views

Reproducible Emu-Based Workflow for High-Fidelity Soil and Plant Microbiome Profiling on HPC Clusters
Henrique M. Dias [...] Christopher Graham
Jan 20, 2026 442 Views
Abstract
The Fusarium genus includes various fungi of great significance in agriculture. Fusarium solani f. sp. eumartii (F. eumartii), traditionally known as a potato pathogen, has also been identified as a cause of disease in tomatoes. This protocol provides a detailed, efficient, and robust flood-inoculation method for assessing F. eumartii infection of young tomato seedlings grown on MS medium plates. It includes the evaluation of the lesion area and the quantification of the remaining fungal inoculum in tomato seedlings. In summary, the straightforwardness and efficiency of this bioassay make it a powerful quantitative tool for selecting fungicidal compounds or defense response inducers in tomato plants, offering a promising approach with significant potential for preventing fungal diseases in crops.
Key features
• Accelerated fungal infection method in tomato seedlings, shortening overall experimental time.
• Allows simultaneous evaluation of fungal infectivity and quantitative remaining inoculum.
• Easily adaptable for screening fungicides and defense inducers in various plant–pathogen interactions.
Keywords: FusariumGraphical overview
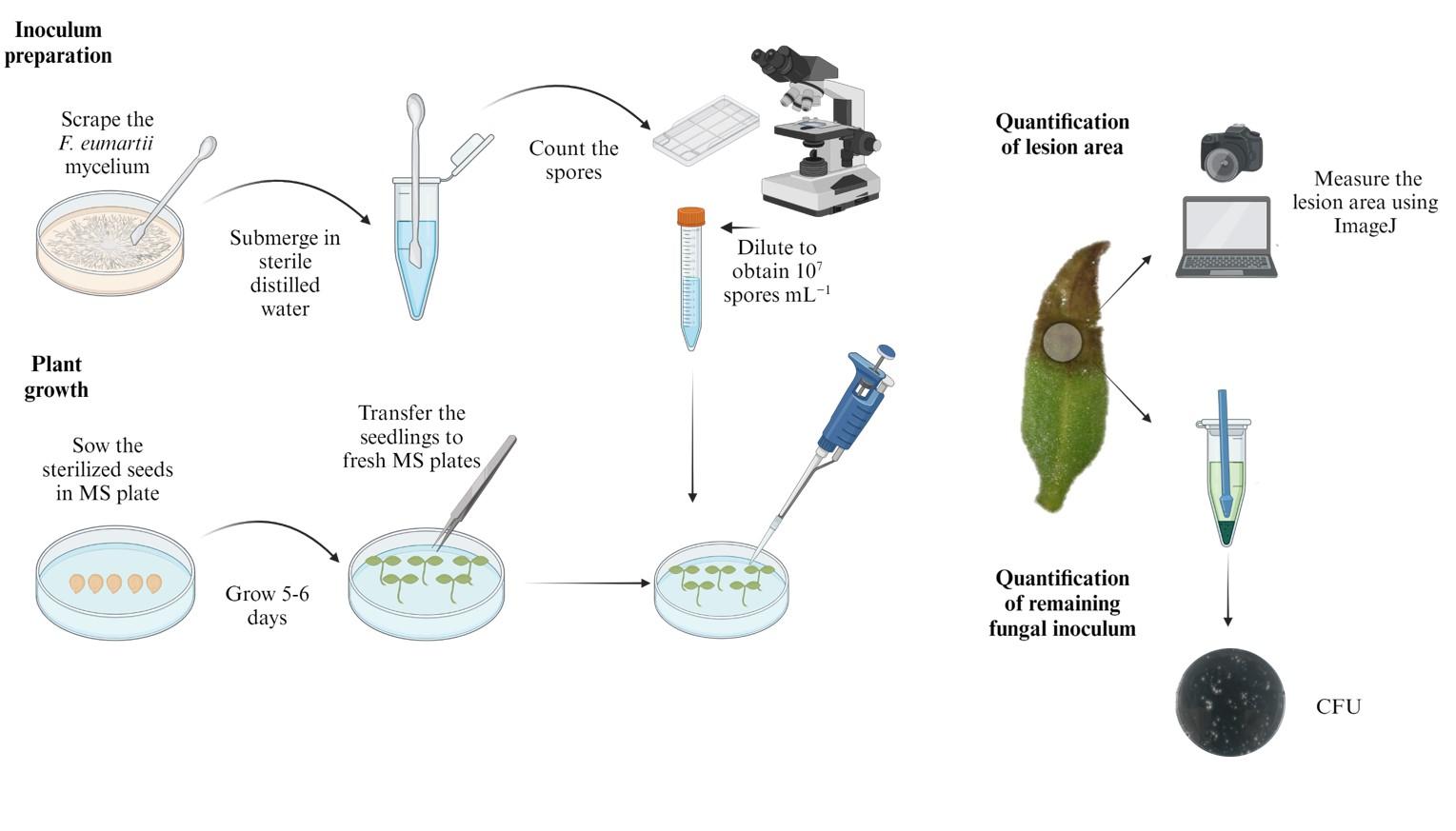
Graphical representation of the protocol for assessing the infectivity of Fusarium solani f. sp. eumartii in tomato seedlings using a flood-inoculation method. The diagram outlines the main steps: i) preparation of the inoculum from fungal mycelium, ii) germination and growth of tomato seedlings, iii) inoculation of seedlings with the spore suspension, iv) infection assessment by measuring lesion areas and remaining fungal inoculum, and v) data analysis. Arrows indicate the sequence of the experimental procedure.
Background
Numerous species within the Fusarium genus, such as Fusarium oxysporum and Fusarium solani, are responsible for significant agricultural losses worldwide, both before and after harvest [1]. Fusarium solani f. sp. eumartii (F. eumartii) is a significant post-harvest disease affecting potato tubers, leading to reddish-brown mottling between leaf veins and dry rot [2]. While traditionally recognized as a potato pathogen (Solanum tuberosum), F. eumartii has also been identified in tomato (Solanum lycopersicum) plants. Isolates from these plant species have also shown pathogenic effects in other members of the Solanaceae family [3]. Different methods of infecting tomato plants with Fusarium fungi have been reported in the literature. In this protocol, we describe a simple, reliable, and reproducible way to assess the infection of tomato seedlings with this pathogen. This method was adapted from a seedling-based assay initially developed by Uppalapati et al. [4] for Pseudomonas syringae pv. tomato, demonstrating its versatility and effectiveness in studying plant-pathogen interactions. Moreover, this approach allows a practical screening of different compounds that can act with fungicides, as described in the work of Terrile et al. [5].
Materials and reagents
Biological materials
1. Fusarium solani f. sp. eumartii (F. eumartii) isolated 3122 (Estación Experimental Agropecuaria -EEA-INTA, Balcarce, Argentina; Radtke and Escande, 1973)
2. Tomato (Solanum lycopersicum cv. Platense) was obtained from a commercial supply
Reagents
1. Agar (BD-DIFCO, catalog number: 214530)
2. Ampicillin (Amp) (GenBiotech, catalog number: A-0104-5)
3. Commercial bleach (sodium hypochlorite <5%)
4. Ethanol (Biopack, catalog number: 2000165400)
5. Murashige and Skoog (MS) medium (Sigma, catalog number: M5524)
6. KOH (Sigma, catalog number: 221473)
7. Potato dextrose agar (PDA) (OXOID, catalog number: CM0139)
8. Silwet L77 (Momentive, catalog number: A0300026)
9. Sterile distilled water
10. Triton X-100 (Biopack, catalog number: 2000200308)
Solutions
1. MS medium, 1 L (see Recipes)
2. PDA medium, 1 L (see Recipes)
3. 30% bleach solution (see Recipes)
4. 70% ethanol (see Recipes)
Recipes
1. MS medium (1 L)
4.4 g of MS basal medium
8 g of agar
Adjust to pH 5.7 with 1 M KOH and autoclave at 121 °C for 15 min
2. PDA medium (1 L)
39 g of PDA
Autoclave at 121 °C for 15 min
3. 30% bleach solution
Add 3 mL of bleach into 7 mL of distilled water and 0.04% Triton X-100
4. 70% ethanol
Add 73 mL of ethanol 96% into 27 mL of distilled water.
Laboratory supplies
1. Forceps
2. Conical tubes (15 and 50 mL) (Deltalab, catalog number: 429942 and 429931)
3. Household aluminum foil for multiple uses
4. 1.5 mL microcentrifuge tubes (Deltalab, catalog number: 4092.4N)
5. Neubauer chamber (Marienfeld, catalog number: 0610101)
6. Paper towels
7. Parafilm (Amcor, catalog number: PM-996)
8. Petri dishes 90 × 14 mm (Deltalab, catalog number: 200200)
9. 0.5 mm diameter punching tool
10. 5 mL pipette tips (Deltalab, catalog number: 900038.C)
11. 1 mL pipette tips (Deltalab, catalog number: 200012)
12. 2–20 μL pipette tips (Deltalab, catalog number: 200016B)
13. Micro-pestle (Axygen, catalog number: AXYPES15BSI)
Equipment
1. Cultivation room at 25 °C under 250 μmol photons m-2 s-1 with 70% ± 5% relative humidity and 16:8 h light/dark cycle
2. Cold room at 4 °C
3. Laminar air flow equipment (CASIBA, model: 3101-FLV-2)
4. Pipettes: PIPETMAN P20L, P200, P1000, P5000 (Gilson, model: F144056M, F144058M, F144059M and F144066)
5. Vortex (VELP Scientifica, model: ZX-3)
6. Stereomicroscope (Nikon, model: SMZ800)
7. Digital camera (Canon, model: DS126431)
Software and datasets
1. ImageJ software (NIH, https://imagej.net/)
2. Graphpad Prism version 5.01 software for Windows (GraphPad Software, La Jolla, California, USA, 2007)
Procedure
A. Fungal growth and inoculum preparation
1. Critical step: Prepare PDA plates supplemented with Amp (100 μg/mL) to prevent bacterial contamination during the growth of F. eumartii while maintaining the viability and functionality of fungi.
2. Cut a 0.5 mm PDA disc with sterile punching tools from plates previously colonized with F. eumartii mycelium.
3. Place the disc in the center of the freshly prepared plate, with the mycelium toward the culture medium (Figure 1A).
4. Incubate the plates in the cultivation room at 25 °C for 6–7 days in darkness. At this time, all the plates are covered with fungal mycelium (Figure 1B).
5. Store the plates at 4 °C in darkness.
6. To prepare inoculum (spore suspension) for the bioassay, scrape the mycelium superficially with a sterile spatula to obtain spores and submerge in a 1.5 mL microcentrifuge tube with 1 mL of sterile distilled water (Figure 1C).
7. Count the spores in a Neubauer chamber.
8. Dilute the original spore suspension to obtain a concentration of 107 spores/mL.
Note: If the action of a fungicidal compound is to be evaluated, treatment and incubation of the spores can be performed at this point before plant infection.
9. Add Silwet L77 to achieve a final concentration of 0.025% (v/v) and mix thoroughly.
Note: Use the fungal suspensions immediately after preparation.
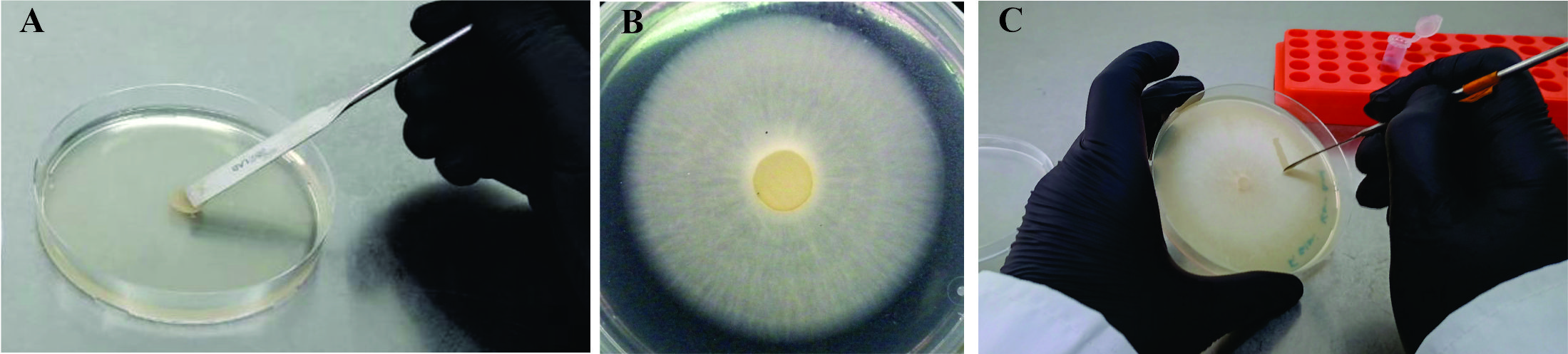
Figure 1. Fungal growth and inoculum preparation. A. Inoculation of freshly prepared PDA plate with fungus-colonized PDA disc. B. 7-day F. eumartii mycelia growth on PDA plate. C. Scraping of mycelium to obtain a spore solution to inoculate plants.
B. Plant growth
1. Seed sterilization
a. Incubate 500 tomato seeds (approximately 1 g) in 10 mL of 70% ethanol for 2 min in a 15 mL conical tube.
b. Discard the ethanol solution by pipetting. Caution: Rinse the seeds twice with sterile distilled water to avoid ethanol residues.
c. Incubate the seeds with 10 mL of 30% bleach solution for 15 min with gentle shaking (500–1,000 rpm).
d. Wash the seeds thoroughly with 10 mL of sterile distilled water three times. Caution: Ensure complete removal of bleach residues to prevent seed damage or interference with germination.
Pause point: The seeds can be kept in sterile distilled water at 4 °C for a few hours before sowing on the MS plates.
2. Seed germination
a. Sow the seeds with sterile forceps in Petri dishes with MS medium (20–30 seeds per plate; Figure 2A).
Note: Prepare at least three plates per treatment for quantitative analysis.
b. Incubate the plates horizontally in the cultivation room at 25 °C for 5–6 days until cotyledons emerge.
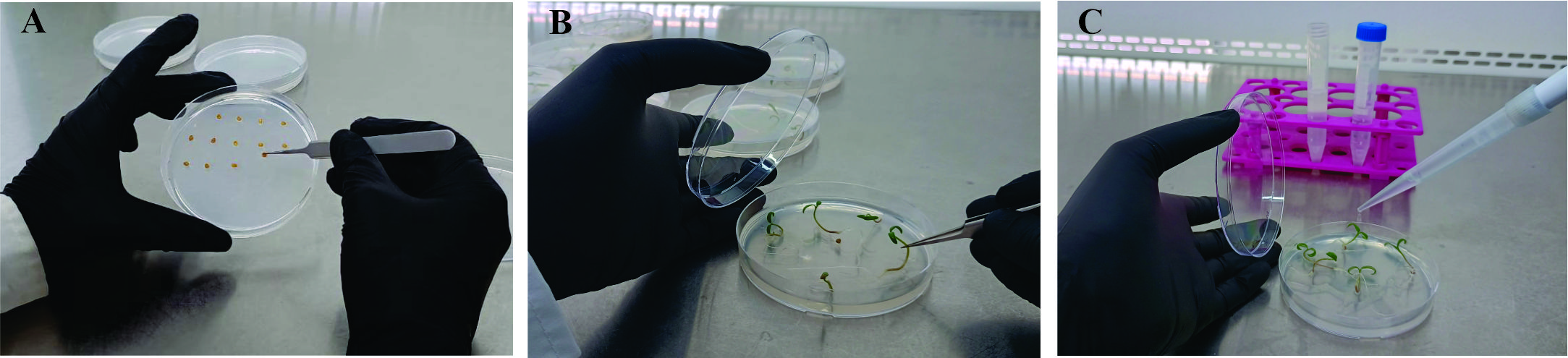
Figure 2. Plant growth and F. eumartii flood inoculation of tomato seedlings. A. Sowing of seed on MS plates. B. Transferring 5-day-old healthy seedlings to fresh MS medium plate. C. Inoculation with spore suspension into cotyledons.
C. In planta bioassay
1. Carefully transfer the healthy seedlings with fully emerged cotyledons to fresh MS medium plates (five seedlings per plate) using sterile forceps (Figure 2B). Ensure that the cotyledons and roots are handled gently to avoid physical damage, which could compromise seedling health or affect infection outcomes.
Note: If desired, transfer the seedlings to MS medium plates supplemented with the treatments to be tested as fungicides.
2. Add 5 mL of the spore suspension to each Petri dish, ensuring that the tomato cotyledons are thoroughly moistened. Incubate for 10–15 min, occasionally mixing the solution to maintain even coverage (Figure 2C). As a control, add 5 mL of a solution containing sterile distilled water and Silwet L77 (at a final concentration of 0.025% v/v) to a separate set of Petri dishes with tomato seedlings.
3. Discard the inoculum suspension and seal the plates with parafilm.
4. Incubate the plates in the cultivation room at 25 °C for 5–6 days until disease symptoms appear.
Caution: To ensure accuracy and reliability, independent sets of cotyledons are used to quantify the lesion area and measure the remaining fungal inoculum. Cotyledons selected for lesion area quantification remain undisturbed for imaging and measurement, while those allocated for inoculum analysis are processed separately to minimize cross-contamination and variability between analyses.
D. Quantification of tomato lesion area
1. Take a photograph of each tomato cotyledon under a stereomicroscope to capture detailed images of the lesion areas (Figure 3A).
Note: To standardize image quality, pictures of tomato cotyledons were taken under consistent lighting conditions using a stereomicroscope equipped with a ring light (intensity set to 75%) to minimize shadows. A digital camera (Canon DS126431) was set to manual mode. Images were saved in JPEG format at maximum resolution.
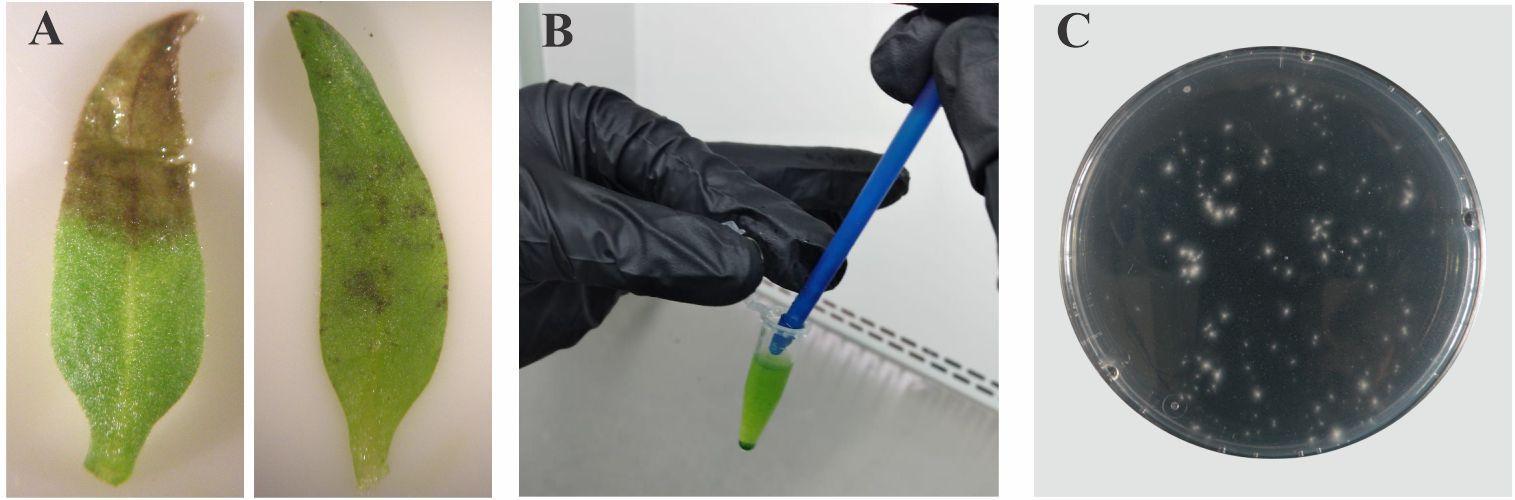
Figure 3. Visual infection symptoms and quantification of remaining fungal inoculum. A. Symptoms of infection in cotyledons (left, pronounced symptoms; right, mild symptoms). B. Homogenizing to effectively recover the remaining inoculum. C. Counting CFU after serial dilutions.
2. Measure the lesion area in the photographed cotyledons using the image-processing software ImageJ. Convert the images to an 8-bit grayscale format to enhance contrast and facilitate measurements. To measure one lesion area (Figure 3A, left cotyledon), use the polygon selection tool to delineate the total area of the cotyledon and select the measure tool (Ctrl+M) to obtain the area value, which will be considered 100%. Repeat the previous sequence by selecting the infected area. Record the area value for the infected region and calculate the lesion area percentage as follows: (infected area/total area) × 100.
3. To analyze multiple lesion areas (Figure 3A, right cotyledon), use the polygon selection tool to delineate each infected region on the cotyledon. After selecting each region, add it to the ROI manager by pressing "Add [t]" in the ROI manager window. Repeat this process for all visible lesions. Then, select the measure tool to obtain the cumulative area of all regions and calculate the lesion area percentage as follows: (cumulative infected area/total area) × 100.
E. Measurement of residual fungal inoculum
1. Remove the cotyledons from the seedlings of each plate, place them in a 15 mL conical tube, and add 10 mL of 70% ethanol for 1 min to eliminate epiphytic bacteria before sampling. Rinse the plant tissue gently with sterile distilled water.
2. Dry the tissue with regular paper towels. Collect approximately 50 mg of the set of cotyledons and place them in a 1.5 mL microcentrifuge tube.
Note: Cotyledons from two independent replicate plants are pooled for a single tissue sample. Three or more samples are needed for each treatment.
3. Add 500 μL of cold sterile distilled water to the tube and homogenize with a plastic micro-pestle by hand until pieces of tissue are no longer visible (Figure 3B). Critical: Ensure the tissue is thoroughly homogenized to prevent discrepancies in CFU counts.
4. Vortex the homogenates for 5 s.
5. Serially dilute the sample in sterile distilled water.
Note: The number of serial dilutions required to obtain countable colonies should be determined empirically for each sample, but dilutions up to 10–7 are usually sufficient.
6. Plate the dilutions in PDA plates supplemented with 100 μg/mL Amp and incubate at 25 °C for 2 days in darkness. Count the CFU using appropriately diluted samples (Figure 3C).
Data analysis
Analyze the data set with Student’s t-test for comparing two averages of fungal growth or ANOVA with post-hoc Tukey’s multiple range test to compare more than two averages of fungal growth.
Validation of protocol
This protocol has been used and validated in the following research article:
Terrile et al. [5]. Nitric oxide–mediated cell death is triggered by chitosan in Fusarium eumartii spores. Pest management science.
The key findings presented in this reference work are summarized as follows:
The validation results highlight the dual effectiveness of chitosan against Fusarium eumartii in tomato seedlings through direct fungicidal action and induced plant defenses. In seedlings inoculated with chitosan-treated spores, necrotic symptoms were reduced by approximately 90%, and residual fungal inoculum decreased by 83% compared to untreated controls. Additionally, pre-treating seedlings with 10 μg/mL of chitosan for 4 days significantly reduced both lesion area and remaining fungal inoculum following spore inoculation. These findings confirm that chitosan not only inhibits fungal growth but also acts as an elicitor of plant defense mechanisms, enhancing the plant's resistance to infection.
General notes and troubleshooting
General notes
The F. eumartii plates used to obtain spore solution should not be more than 3–4 weeks old because older plates may have reduced spore viability or altered fungal physiology/pathogenicity. This could impact the consistency and reliability of the inoculum, leading to variability in infection outcomes and potentially compromising the experiment's reproducibility and accuracy.
Troubleshooting
Problem 1: Absence of disease symptoms on tomato seedlings after 5–6 days post-inoculation.
Possible causes: The inoculum may not be infective, the inoculation may not have been efficient, or the environmental conditions in the growth chamber may not be suitable for infection.
Solution: Ensure that the inoculum is applied uniformly to the cotyledons. If symptoms are not observed after 5–6 days, extend the incubation period and monitor the cultivation room parameters (temperature, humidity, and light). To verify the viability of the inoculum used for inoculation, incubate spore suspension on multi-well microscope slides for 16 h at 25 °C and 100% humidity in darkness. Check for germination under a microscope. Germination should exceed 80% for viability.
Problem 2: Inconsistent quantification of fungal population in inoculated plant tissue.
Possible causes: Variability in the effectiveness of tissue homogenization, inconsistencies in sample preparation methods, or unintentional contamination during handling.
Solution: Use a precise weight of plant tissue for each sample, ensuring uniformity across replicates and treatments. Homogenize the tissue thoroughly to achieve a consistent suspension. Perform dilutions consistently using a fixed volume of homogenized tissue extract for initial dilution and use calibrated pipettes to ensure accuracy in volumes during dilution steps. Make positive control, including a known concentration of fungal spores to validate the dilution and plating process. Additionally, perform parallel quantification of CFU from an uninfected tissue sample as a negative control to account for contamination.
Acknowledgments
This work was partially supported by grants from Agencia Nacional de Promoción de la Investigación, el Desarrollo y la Innovación (Agencia I+D+i, PICT2019-3002 and PICT Start Up 0023), Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET, PIP 0237); Fundación Williams and Universidad Nacional de Mar del Plata (EXA1137/23).
Competing interests
The authors declare that they have no conflict of interest.
References
- K. Chehri. (2011). Occurrence of Fusarium species associated with economically important agricultural crops in Iran. Afr J Microbiol Res. 5(24): e158. https://doi.org/10.5897/ajmr10.158
- D’Ippólito, S., Martín, M. L., Salcedo, M. F., Atencio, H. M., Casalongué, C. A., Godoy, A. V. and Fiol, D. F. (2010). Transcriptome profiling of Fusarium solani f. sp. eumartii -infected potato tubers provides evidence of an inducible defense response. Physiol Mol Plant Pathol. 75: 3–12. https://doi.org/10.1016/j.pmpp.2010.09.002
- Romberg, M. K. and Davis, R. M. (2007). Host Range and Phylogeny of Fusarium solani f. sp. eumartii from Potato and Tomato in California. Plant Dis. 91(5): 585–592. https://doi.org/10.1094/pdis-91-5-0585
- Uppalapati, S. R., Ishiga, Y., Wangdi, T., Urbanczyk-Wochniak, E., Ishiga, T., Mysore, K. S. and Bender, C. L. (2008). Pathogenicity of Pseudomonas syringae pv. tomato on Tomato Seedlings: Phenotypic and Gene Expression Analyses of the Virulence Function of Coronatine. Mol Plant-Microbe Interact. 21(4): 383–395. https://doi.org/10.1094/mpmi-21-4-0383
- Terrile, M. C., Mansilla, A. Y., Albertengo, L., Rodríguez, M. S. and Casalongué, C. A. (2014). Nitric‐oxide‐mediated cell death is triggered by chitosan in Fusarium eumartii spores. Pest Manage Sci. 71(5): 668–674. https://doi.org/10.1002/ps.3814
Article Information
Publication history
Received: Sep 25, 2024
Accepted: Dec 15, 2024
Available online: Jan 5, 2025
Published: Feb 5, 2025
Copyright
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Terrile, M. C., Mesas, F. A., Picco, M. E., Salcedo, M. F. and Mansilla, A. Y. (2025). Flood Inoculation of Fusarium eumartii in Tomato Seedlings: Method for Evaluating the Infectivity of Pathogen Spores. Bio-protocol 15(3): e5198. DOI: 10.21769/BioProtoc.5198.
Category
Plant Science > Plant immunity > Host-microbe interactions
Microbiology > Microbe-host interactions > Fungus
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









