- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Campylobacter jejuni Biofilm Assessment by NanoLuc Luciferase Assay
Published: Vol 15, Iss 4, Feb 20, 2025 DOI: 10.21769/BioProtoc.5192 Views: 1613
Reviewed by: Emilia KrypotouAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Leishmania Parasite Quantification by Bioluminescence in Murine Models
Marta Reverte and Nicolas Fasel
Nov 20, 2019 4676 Views
Listeria innocua Biofilm Assay Using NanoLuc Luciferase
Aleš Berlec [...] Jerica Sabotič
Feb 5, 2022 3350 Views
Abstract
Campylobacter jejuni, a widespread pathogen found in birds and mammals, poses a significant risk for zoonosis worldwide despite its susceptibility to environmental and food-processing stressors. One of its main survival mechanisms is the formation of biofilms that can withstand various food-processing stressors, which is why efficient methods for assessing biofilms are crucial. Existing methods, including the classical culture-based plate counting method, biomass-staining methods (e.g., crystal violet and safranin), DNA-staining methods, those that use metabolic substrates to detect live bacteria (e.g., tetrazolium salts and resazurin), immunofluorescence with flow cytometry or fluorescence microscopy, and PCR-based methods for quantification of bacterial DNA, are diverse but often lack specificity, sensitivity, and suitability. In response to these limitations, we propose an innovative approach using NanoLuc as a reporter protein. The established protocol involves growing biofilms in microtiter plates, washing unattached cells, adding Nano-Glo luciferase substrate, and measuring bioluminescence. The bacterial concentrations in the biofilms are calculated by linear regression based on the calibration curve generated with known cell concentrations. The NanoLuc protein offers a number of advantages, such as its small size, temperature stability, and highly efficient bioluminescence, enabling rapid, non-invasive, and comprehensive assessment of biofilms together with quantification of a wide range of cell states. Although this method is limited to laboratory use due to the involvement of genetically modified organisms, it provides valuable insights into C. jejuni biofilm dynamics that could indirectly help in the development of improved food safety measures.
Key features
• Quantification of C. jejuni using NanoLuc luciferase.
• The assay is linear in the range of 1.9 × 107 to 1.5 × 108 CFU/mL.
• Following biofilm growth, less than 1 h is required for detection.
• The assay requires genetically modified bacterial strains.
Keywords: Campylobacter jejuniGraphical overview
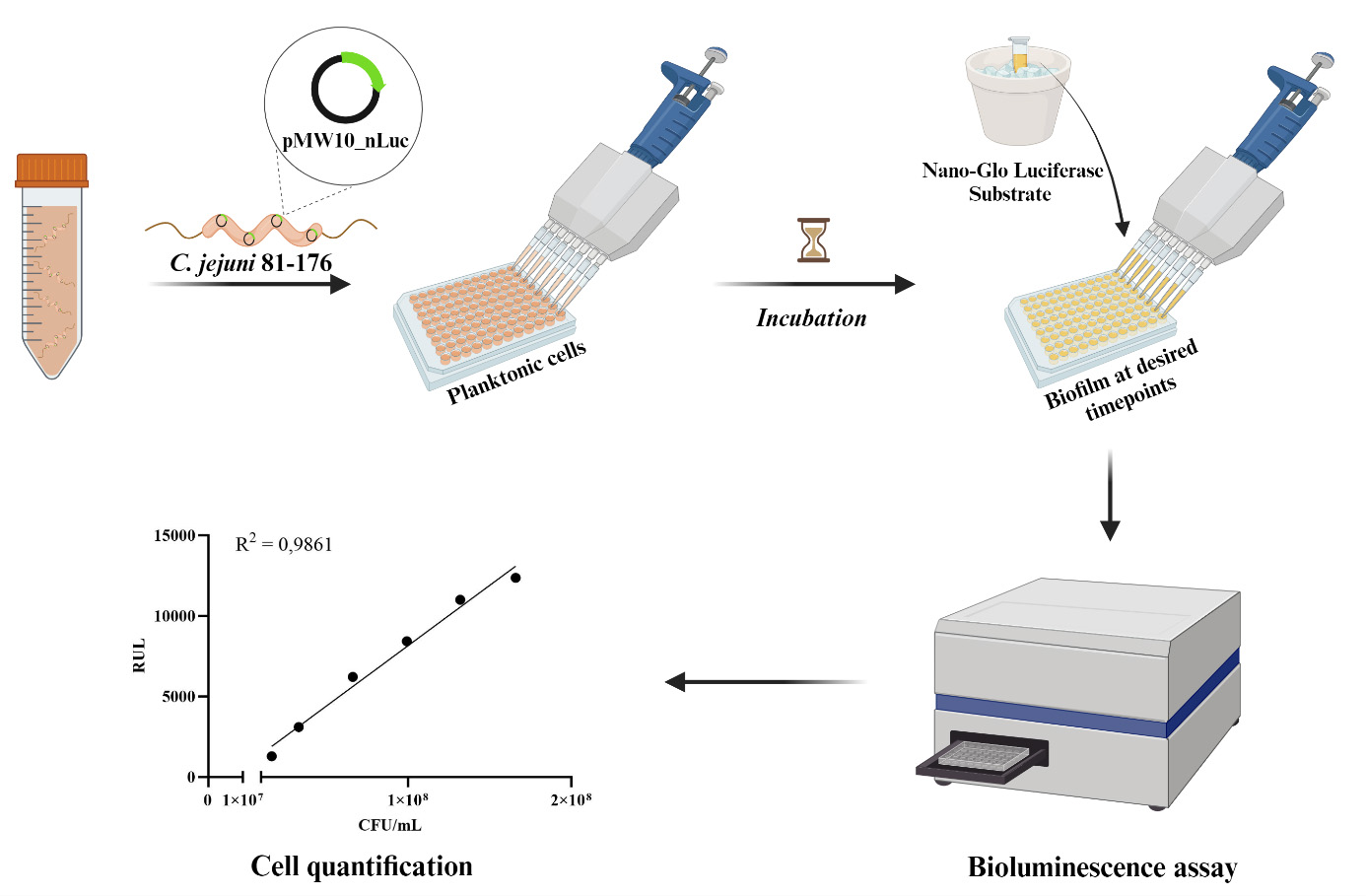
Campylobacter jejuni biofilm detection using NanoLuc luciferase assay
Background
Campylobacter jejuni, a pathogen found in the gastrointestinal tract of both domestic and wild birds and mammals, is a major cause of zoonosis worldwide [1]. However, it is sensitive to environmental stressors outside its hosts. To this end, C. jejuni forms biofilms as a survival strategy. Biofilms are characterized by a unique bacterial growth state and act as a protective shield that enables bacterial cells to resist certain environmental stressors—such as temperature fluctuations, desiccation, pH changes, and oxidative stress—commonly found on food preparation surfaces, food products, and water reservoirs [2–5]. Bacteria adapt to environmental stressors by, for example, adopting the nonmotile coccoid form and entering a viable but nonculturable state (VBNC) [6–8]. Viable but nonculturable cells cannot divide in vitro but maintain their membrane integrity and metabolic activity [9], which hinders their detection in quality control tests (a critical aspect of food safety) and may explain the persistence of campylobacteriosis in developed countries despite strict hygiene standards [1]. Studies suggest that bacteria in the VBNC state not only remain viable but also have the potential to recover and cause infections [10,11]. To assess biofilms, conventional cultivation methods and colony counts are generally used and usually complemented with biomass estimation using crystal violet staining [12,13]. The metabolic activity of biofilms can be assessed colorimetrically by monitoring the conversion of tetrazolium salt to formazan [14]. Simultaneously, ATP can be measured using BacTiter-Glo or resazurin fluorescence [8,12,13]. Advanced techniques such as transmission electron microscopy, scanning electron microscopy, and immunofluorescence with flow cytometry or fluorescence confocal laser scanning microscopy provide detailed information on the heterogeneity, cell localization, and structure of biofilms [15–17]. Additionally, PCR-based methods are used [12]. However, most of these methods are expensive, unspecific, technically demanding, and unsuitable for cell quantification. Therefore, new methods that can efficiently and rapidly detect C. jejuni cells would be valuable for advancing research related to food safety and possible applications in the food industry. The reporter protein NanoLuc offers several advantages, such as small size (19 kDa), high temperature stability (Tm = 60 °C), activity over a broad pH range (6–8), and the absence of post-translational modifications or disulfide bonds, which ensures uniform distribution within cells. Its most outstanding feature is its highly efficient bioluminescence. The reaction involves the NanoLuc enzyme, which is constitutively expressed, and the Nano-Glo substrate, commonly referred to as luciferin. Upon addition of the substrate in the presence of oxygen, NanoLuc (as luciferase) catalyzes the oxidation of luciferin and generates oxyluciferin. This molecule releases light in its excited energy state when it returns to its ground state [18,19]. No ATP is required for the luminescence reaction, which helps to minimize background luminescence. This reduction in the background signal ensures that the luminescence signal more accurately reflects the metabolic activity of the cells without external interference [20]. NanoLuc has already been successfully used to monitor and quantify Listeria innocua biofilms [21], and we have adapted the protocol to assess C. jejuni growth and biofilm formation. The protocol provides a comprehensive approach to biofilm monitoring and can be used as an important complement or alternative to conventional biofilm monitoring methods.
Materials and reagents
Biological materials
1. C. jejuni 81-176 (GenBank accession number NC_008787.1) transformed with pMW10_nLuc (GenBank accession number OR958835) (Figure 1)
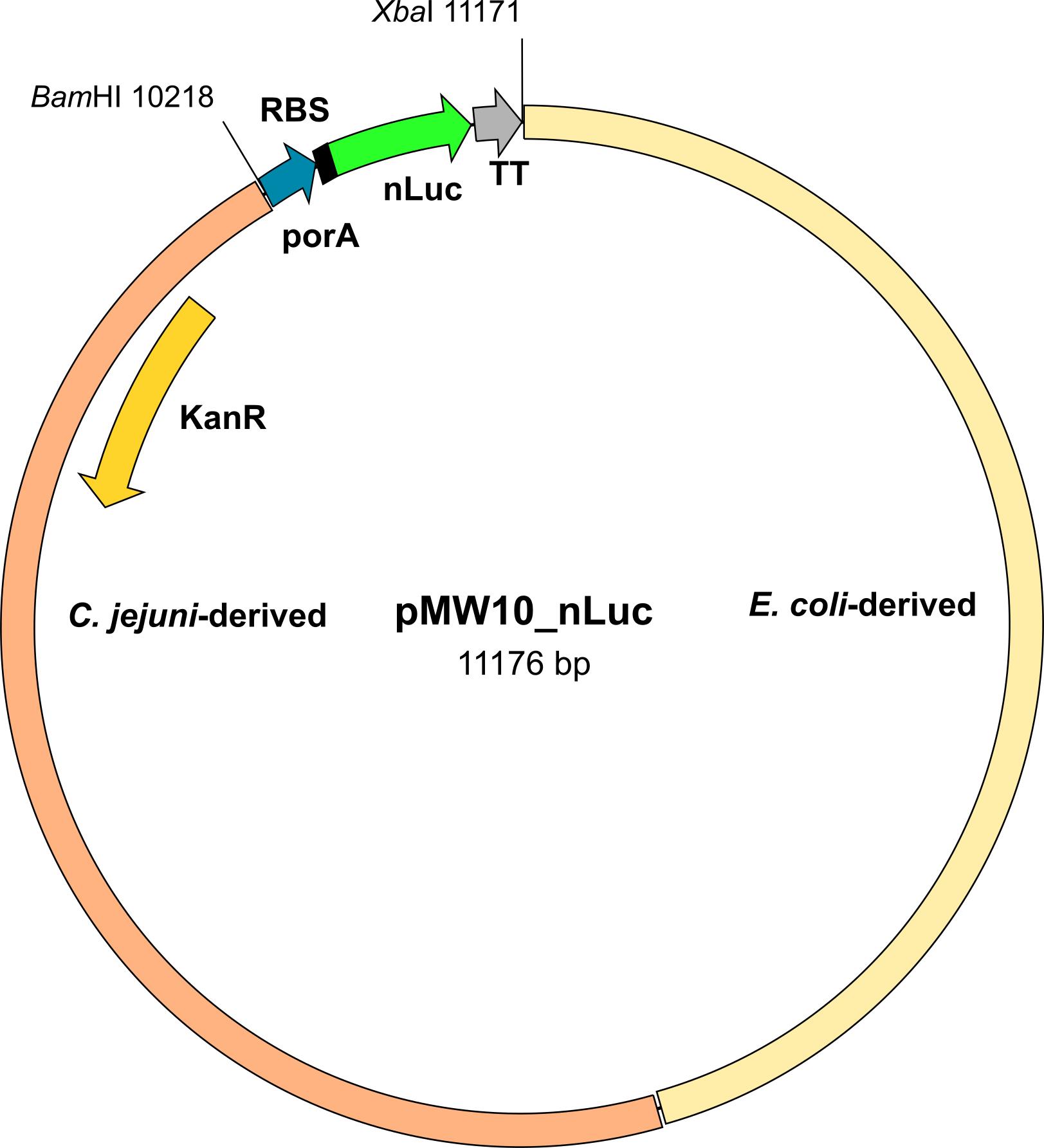
Figure 1. Scheme of the pMW10_nLuc plasmid. KanR, kanamycin resistance gene; porA, promotor; RBS, ribosome-binding site; nLuc, NanoLuc gene; TT, transcription terminator; BamHI and XbaI, restriction sites.
Reagents
1. Nano-Glo Luciferase Assay System (Promega, catalog number: N1120)
2. Phosphate-buffered saline (PBS) (Sigma-Aldrich, catalog number: P3813)
3. Mueller-Hinton broth (MHB) powder (BD Difco, catalog number: 275730)
4. Mueller-Hinton agar (MHA) powder (BD Difco, catalog number: 225250)
5. Karmali Campylobacter selective supplement (Oxoid, catalog number: SR0167E)
6. Ethanol, 99.8%, for HPLC, absolute (Thermo Scientific, catalog number: 445740010)
7. Kanamycin sulfate from Streptomyces kanamyceticus (BioReagent, Sigma-Aldrich, K1377)
Solutions
1. PBS solution (see Recipes)
2. MHB medium/MHA agar (see Recipes)
3. MHA Karmali agar (see Recipes)
Recipes
1. PBS solution
One pouch of PBS
1 L of purified water
Mix thoroughly. Autoclave at 121 °C for 15 min. Avoid overheating.
2. MHB medium/MHA agar
38 g of MHB powder or 21 g of MHA powder
1 L of purified water
Mix thoroughly. Heat with frequent agitation and boil for 1 min to completely dissolve the MHB or MHA powder. Autoclave at 121 °C for 15 min. Avoid overheating. Pour the MHA agar into sterile Petri dishes.
3. MHA Karmali agar
19 g of MHA powder
500 mL of purified water
One vial of Karmali Campylobacter selective supplement
1 mL of sterile distilled water
1 mL of 99.8% ethanol
Aseptically add 2 mL ethanol/sterile distilled water in a 1:1 ratio to the vial containing the Karmali Campylobacter selective supplement and mix gently to ensure that the contents dissolve completely. Add to 500 mL of the prepared MHA agar cooled to 50 °C (see Recipe 2). Mix well and pour into sterile Petri dishes.
Laboratory supplies
1. White flat-bottomed 96-well plates (Thermo Fisher, Microlite, catalog number: 7417)
2. 50 mL centrifuge tube (Corning, Mini Bioreactor, catalog number: 431720)
3. 1.5 mL microcentrifuge tubes (Eppendorf, Safe-Lock, catalog number: EP0030123611)
4. Pre-sterile 50 mL disposable reservoirs (Biotix, catalog number: EP0030123611)
5. Microplate sealing tape (Corning, catalog number: 3345)
6. Pipette tips 0.1–10 μL (Eppendorf, catalog number: 0030000811), 2–200 μL (Eppendorf, catalog number: 0030000889), 50–1,000 μL (Eppendorf, catalog number: 0030000927)
7. Inoculation loops (Golias, catalog number: EZ08)
Equipment
1. Incubator (Thermo Scientific)
2. Orbital shaker (Thermo Scientific)
3. Luminescence plate reader (BMG LabTech, FLUOstar Omega)
4. Pipette set (Eppendorf, catalog number: 05-403-151)
5. Multichannel pipette (Eppendorf, model: Research Plus, catalog number: 2231300048)
6. Biosafety cabinet (Nuaire)
7. Deep freezer (Haier, model: TwinCool)
8. Spectrophotometer (Bio-Rad, model: SmartSpec 3000, catalog number: 4006168)
9. Centrifuge (Thermo Scientific, model: LEGEND X1R, catalog number: 75004261)
Procedure
A. Reviving glycerol stock of C. jejuni
1. Remove a vial containing a stock culture of C. jejuni, previously transformed with the pMW10_nLuc plasmid via triparental mating [22], from -80 °C and place it on ice. For handling C. jejuni, see General note 1.
2. Using a sterile loop, aseptically transfer half a loop of culture to Karmali-selective plates and incubate the plates at 42 °C under microaerophilic conditions (85% N2, 10% CO2, and 5% O2) for 24 h. Microaerophilic conditions are ensured by blowing the gas mixture into an airtight box, which is then placed in the incubator.
3. Using a cotton swab, respread the colonies grown on Karmali agar to MHA agar containing 30 μg/mL of kanamycin. Incubate the plate at 42 °C under microaerophilic conditions for 24 h.
B. Overnight culture for calibration curve preparation
1. Prepare 15 mL of MHB medium with 30 μg/mL of kanamycin in a 50 mL centrifuge tube.
2. Using a sterile inoculation loop, inoculate a few colonies from the MHA agar plate into the medium prepared above. Vortex the tubes.
3. Incubate at 42 °C under microaerophilic conditions (85% N2, 10% CO2, and 5% O2) with gentle shaking (80 rpm) for 16–20 h.
C. Establishing biofilms in microtiter plates
1. Using a sterile loop, take a few colonies from the MHA agar plate and resuspend them in MHB medium until they reach OD600 = 0.1 (approximately 107 CFU/mL, see General note 2).
2. Transfer 100 μL of this suspension into 9.9 mL of MHB medium to achieve a final concentration of approximately 10 CFU/mL. This will serve as the initial culture.
3. Pipette 200 μL of the initial culture to each well, using three separately prepared samples (biological replicates) with each biological replicate in three wells (technical replicates). This results in nine wells per condition. Cover the plates with sterile microplate sealing tape.
4. Incubate the plates under microaerophilic conditions (85% N2, 10% CO2, and 5% O2) for different time periods (e.g., 4, 8, 24, 48, or 72 h).
5. After incubation, carefully rinse the biofilms in the microtiter plates three times with 100 μL of PBS. After the last wash, add 50 μL of PBS to each well. Proceed with bioluminescence measurements as described in section D. Thaw the Nano-Glo luciferase buffer (from Nano-Glo Luciferase Assay System) and gently mix the Nano-Glo luciferase substrate by pipetting. Allow both the reagents and the cultures in the microtiter plates to equilibrate to room temperature before proceeding with the experiment.
D. Calibration curve preparation
1. Adjust the overnight culture (prepared in section B) using sterile MHB medium to obtain an OD600 corresponding to 1.50 × 108 CFU/mL (see General note 2). This will be the bacterial output suspension for the calibration curve.
2. Prepare five distinct dilutions (0.8, 0.6, 0.4, 0.2, 0.1) in 1.5 mL tubes to create a calibration curve using the prepared bacterial output suspension (1.50 × 108 CFU/mL) as the starting concentration (1.0 or 100%). Prepare each dilution by adding the appropriate volumes of bacterial suspension and sterile growth medium, e.g., tube 0.8: mix 800 μL of bacterial suspension with 200 μL of sterile growth medium, etc.
3. Transfer 50 μL of dilutions to the unused wells of the microtiter plate with biofilms in duplicate.
4. Use sterile MHB medium as a blank, which is placed in three wells. The mean value is subtracted from each measurement.
E. Bioluminescence measurement
1. Thaw the Nano-Glo luciferase buffer and gently mix the Nano-Glo luciferase substrate by pipetting. Allow both the reagents and the cultures in the microtiter plates to equilibrate to room temperature before proceeding with the experiment.
2. Prepare the appropriate amount of substrate by mixing one volume (e.g., 100 μL) of Nano-Glo luciferase assay substrate with 50 volumes (e.g., 5 mL) of Nano-Glo luciferase assay buffer.
3. Quickly add 50 μL of substrate to each well containing the culture and wait 3 min. Then, measure bioluminescence using the plate reader (e.g., Omega FLUOstar) (settings: luminescence mode; integration time, 1 s; settle time, 150 ms).
F. Calculating the concentration of bacteria in biofilms
1. Use Excel to plot the calibration curve with relative light units on the y-axis and CFU/mL on the x-axis. A representative calibration curve is shown in Figure 2.
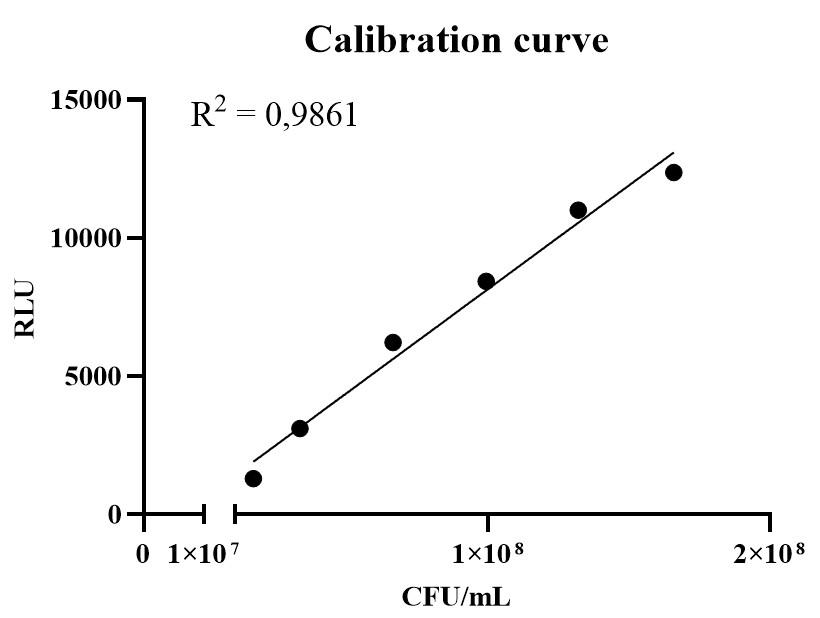
Figure 2. Representative calibration curve of the Campylobacter jejuni biofilm assay. RLU, relative light units; CFU/mL, colony-forming units per mL.
2. Use linear regression (Excel functions: slope and intercept) to calculate biofilm cell concentrations from measured bioluminescence (see General note 3).
Data analysis
Bioluminescence measurements are saved in Excel format, and Excel can be used to analyze the data. The coefficient of determination (R2) of the calibration curve is determined, and data are considered for analysis only if R2 > 0.95. All measurements are repeated in three biological replicates, each of which is repeated in three technical replicates.
Example for the calculation of the biofilm cell concentration based on the measured bioluminescence (RLU): To calculate biofilm cell concentration from RLU, first create a calibration curve using known CFU/mL dilutions (see section F). For example, the curve equation is y = 0.0002x + 36.49 (k = 0.0002; n = 36.49; x = CFU/mL value; y = RLU value). By rearranging the equation, the CFU/mL is calculated from the RLU value of the samples as in Table 1.
Table 1. CFU/mL calculated from the RLU value of the samples
| Sample number | RLU | CFU/mL |
|---|---|---|
| 1 | 11,425 | 5.69 × 107 |
| 2 | 8,054 | 4.01 × 107 |
| 3 | 4,983 | 2.47 × 107 |
Validation of protocol
This protocol was validated in Čukajne et al. [23], DOI: 10.1007/s00253-024-13383-0.
General notes and troubleshooting
General notes
1. Biosafety level 2 (BSL-2) practices must be followed when handling Campylobacter jejuni, which is classified as a Risk Group 2 pathogen. This includes the use of personal protective equipment such as lab coats and gloves, working in Class II biosafety cabinets to prevent exposure to infectious aerosols, and strictly limiting access to the laboratory to trained personnel. Decontamination of biological waste must be ensured prior to disposal, usually by autoclaving [24].
2. The relationship between OD600 and CFU number should be established in advance by plating bacterial suspensions with different OD600 values (serial dilutions) and counting the colonies. Measured OD600 values depend on the spectrophotometer and other experimental conditions used and are therefore not specified. In our settings, for example, OD600 = 1 corresponds to ~1.50 × 108 CFU/mL.
3. In time-course experiments of biofilm growth, each time point (or each plate measured) requires a new calibration curve prepared from the overnight culture. Therefore, an overnight culture should be prepared one day in advance. By including a calibration curve at each time point, we can account for any variations between plates, ensuring accurate measurements despite the high sensitivity of the method.
Acknowledgments
This study was funded by the Slovenian Research Agency (grant numbers J4-3088, J4-4548, J7-4420, P4-0116, and BI-US/22-24-073). This protocol was derived from Čukajne et al. [23], DOI: 10.1007/s00253-024-13383-0. The authors acknowledge Dr. Eva Lasic for editing and reviewing the manuscript.
Competing interests
The authors declare no competing interests.
References
- EFSA, ECDC. (2023). The European Union One Health 2022 Zoonoses Report. EFSA J. 21(12): e8442.
- Tram, G., Day, C. J. and Korolik, V. (2020). Bridging the Gap: A Role for Campylobacter jejuni Biofilms. Microorganisms. 8(3): 452.
- Joshua, G. W. P., Guthrie-Irons, C., Karlyshev, A. V. and Wren, B. W. (2006). Biofilm formation in Campylobacter jejuni. Microbiology. 152(2): 387–396.
- Klančnik, A., Vučković, D., Plankl, M., Abram, M. and Smole Možina, S. (2013). In Vivo Modulation of Campylobacter jejuni Virulence in Response to Environmental Stress. Foodborne Pathog Dis. 10(6): 566–572.
- Klančnik, A., Vučković, D., Jamnik, P., Abram, M. and Smole Možina, S. (2014). Stress Response and Virulence of Heat-Stressed Campylobacter jejuni. Microbes Environ. 29(4): 338–345.
- Tangwatcharin, P., Chanthachum, S., Khopaibool, P. and Griffiths, M. W. (2006). Morphological and Physiological Responses of Campylobacter jejuni to Stress. J Food Prot. 69(11): 2747–2753.
- Klančnik, A., Zorman, T., and Smole Možina, S. (2008). Effects of Low Temperature, Starvation, and Oxidative Stress on the Physiology of Campylobacter jejuni Cells. Croatica Chemica Acta. 81(1): 41–46.
- Klančnik, A., Šimunović, K., Sterniša, M., Ramić, D., Smole Možina, S. and Bucar, F. (2021). Anti-adhesion activity of phytochemicals to prevent Campylobacter jejuni biofilm formation on abiotic surfaces. Phytochem Rev. 20(1): 55–84.
- Ramamurthy, T., Ghosh, A., Pazhani, G. P. and Shinoda, S. (2014). Current Perspectives on Viable but Non-Culturable (VBNC) Pathogenic Bacteria. Front Public Health. 2: e00103.
- Talibart, R., Denis, M., Castillo, A., Cappelier, J. and Ermel, G. (2000). Survival and recovery of viable but noncultivable forms of Campylobacter in aqueous microcosm. Int J Food Microbiol. 55: 263–267.
- Baffone, W., Casaroli, A., Citterio, B., Pierfelici, L., Campana, R., Vittoria, E., Guaglianone, E. and Donelli, G. (2006). Campylobacter jejuni loss of culturability in aqueous microcosms and ability to resuscitate in a mouse model. Int J Food Microbiol. 107(1): 83–91.
- Klančnik, A., Šikić Pogačar, M., Trošt, K., Tušek Žnidarič, M., Mozetič Vodopivec, B. and Smole Možina, S. (2017). Anti-Campylobacter activity of resveratrol and an extract from waste Pinot noir grape skins and seeds, and resistance of Camp. jejuni planktonic and biofilm cells, mediated via the CmeABC efflux pump. J Appl Microbiol. 122(1): 65–77.
- Trošt, K., Klančnik, A., Mozetič Vodopivec, B., Sternad Lemut, M., Jug Novšak, K., Raspor, P. and Smole Možina, S. (2016). Polyphenol, antioxidant and antimicrobial potential of six different white and red wine grape processing leftovers. J Sci Food Agric. 96(14): 4809–4820.
- Ma, L., Feng, J., Zhang, J. and Lu, X. (2022). Campylobacter biofilms. Microbiol Res. 264: 127149.
- Oh, E., Andrews, K. J. and Jeon, B. (2018). Enhanced Biofilm Formation by Ferrous and Ferric Iron Through Oxidative Stress in Campylobacter jejuni. Front Microbiol. 9: e01204.
- Wagle, B. R., Upadhyay, A., Upadhyaya, I., Shrestha, S., Arsi, K., Liyanage, R., Venkitanarayanan, K., Donoghue, D. J. and Donoghue, A. M. (2019). Trans-Cinnamaldehyde, Eugenol and Carvacrol Reduce Campylobacter jejuni Biofilms and Modulate Expression of Select Genes and Proteins. Front Microbiol. 10: e01837.
- Ramić, D., Bucar, F., Kunej, U., Dogša, I., Klančnik, A. and Smole Možina, S. (2021). Antibiofilm Potential of Lavandula Preparations against Campylobacter jejuni. Appl Environ Microbiol. 87(19): e01099–21.
- Kaskova, Z. M., Tsarkova, A. S. and Yampolsky, I. V. (2016). 1001 lights: luciferins, luciferases, their mechanisms of action and applications in chemical analysis, biology and medicine. Chem Soc Rev. 45(21): 6048–6077.
- Promega Coorporation. (2022). Nano-Glo® Luciferase Assay System Technical Manual.
- England, C. G., Ehlerding, E. B. and Cai, W. (2016). NanoLuc: A Small Luciferase Is Brightening Up the Field of Bioluminescence. Bioconjugate Chem. 27(5): 1175–1187.
- Berlec, A., Janež, N., Sterniša, M., Klančnik, A. and Sabotič, J. (2022). Listeria innocua Biofilm Assay Using NanoLuc Luciferase. Bio Protoc. 12(3): e4308.
- Miller, W. G., Bates, A. H., Horn, S. T., Brandl, M. T., Wachtel, M. R. and Mandrell, R. E. (2000). Detection on Surfaces and in Caco-2 Cells of Campylobacter jejuni Cells Transformed with New gfp, yfp, and cfp Marker Plasmids. Appl Environ Microbiol. 66(12): 5426–5436.
- Čukajne, T., Štravs, P., Sahin, O., Zhang, Q., Berlec, A. and Klančnik, A. (2024). Holistic monitoring of Campylobacter jejuni biofilms with NanoLuc bioluminescence. Appl Microbiol Biotechnol. 108(1): 546.
- Public Health Agency of Canada. (2024). Campylobacter jejuni - Pathogen safety data sheet. Government of Canada. Retrieved November 29, 2024, from https://www.canada.ca/en/public-health/services/laboratory-biosafety-biosecurity/pathogen-safety-data-sheets-risk-assessment/campylobacter-jejuni.html
Article Information
Publication history
Received: Sep 23, 2024
Accepted: Dec 12, 2024
Available online: Jan 8, 2025
Published: Feb 20, 2025
Copyright
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Čukajne, T., Štravs, P., Sahin, O., Zhang, Q., Berlec, A. and Klančnik, A. (2025). Campylobacter jejuni Biofilm Assessment by NanoLuc Luciferase Assay. Bio-protocol 15(4): e5192. DOI: 10.21769/BioProtoc.5192.
Category
Microbiology > Microbial biofilm
Microbiology > Pathogen detection > Bioluminescence
Environmental science > Bacterium
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









