- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Development and Validation of Chlamydia muridarum Mouse Models for Studying Genital Tract Infection Pathogenesis
Published: Vol 15, Iss 3, Feb 5, 2025 DOI: 10.21769/BioProtoc.5181 Views: 1289
Reviewed by: Andrea GramaticaPooja MukherjeeNidhi MenonJohn P Phelan

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Eimeria vermiformis Infection Model of Murine Small Intestine
Patrícia Figueiredo-Campos [...] Marc Veldhoen
Dec 20, 2018 6603 Views
Isolation of GFP-expressing Malarial Hypnozoites by Flow Cytometry Cell Sorting
Annemarie Voorberg-van der Wel [...] Clemens H. M. Kocken
May 5, 2021 4628 Views
Babesia duncani in Culture and in Mouse (ICIM) Model for the Advancement of Babesia Biology, Pathogenesis and Therapy
Vandana Kumari [...] Choukri Ben Mamoun
Nov 20, 2022 1917 Views
Abstract
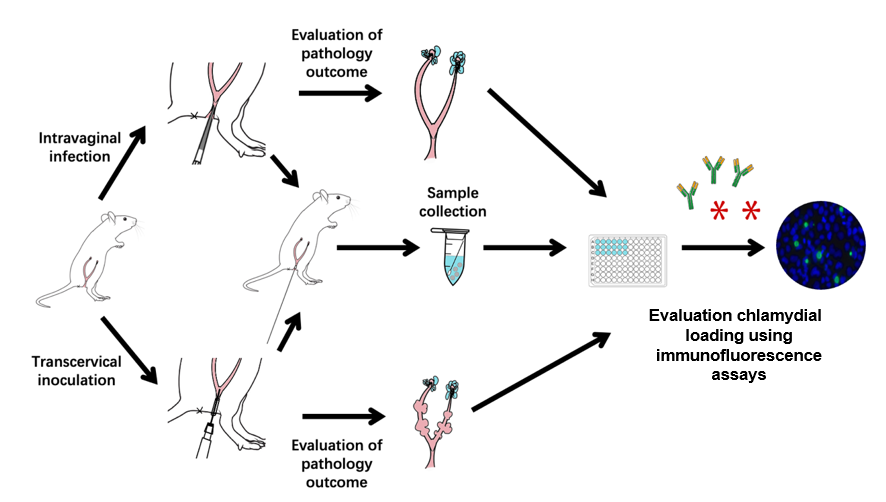
Animal infection models play significant roles in the study of bacterial pathogenic mechanisms and host–pathogen interactions, as well as in evaluating drug and vaccine efficacies. Chlamydia trachomatis is responsible for infections in various mucosal tissues, including the eyes and urogenital, respiratory, and gastrointestinal tracts. Chronic infections can result in severe consequences such as trachoma-induced blindness, ectopic pregnancy, and infertility. While intravaginal inoculation of C. muridarum mimics the natural route of sexual transmission between individuals, transcervical inoculation allows the organisms to directly infect endometrial epithelial cells without interference from host responses triggered by chlamydial contact or infection of vaginal and cervical cells. Therefore, in this study, we used mouse models to visualize pathologies in both the endometrium and oviduct following C. muridarum inoculation.
Key features
• This protocol develops the mouse-adapted Chlamydia muridarum model, ideal for visualizing pathologies in both the endometrium and oviduct genital tract.
• Requires female mice and utilizes specific techniques for intravaginal and transcervical inoculation with chlamydial elementary body (EB) and a form specialized for intracellular replication.
• The protocol necessitates specialized equipment, including a laminar flow hood, a micropipette, and a non-surgical embryo transfer device (NSET).
Keywords: Chlamydia muridarumGraphical overview
Background
All chlamydial species are obligate intracellular bacteria that replicate exclusively inside eukaryotic host cells. Chlamydial infections are dependent on a complex infection cycle that depends on transitions between specific forms. This cycle consists of a cell form specialized for host cell invasion, the elementary body (EB), and a form specialized for intracellular replication, the reticulate body (RB). Chlamydia trachomatis is a leading cause of sexually transmitted bacterial infections. Although C. trachomatis is sensitive to antibiotics, many infected individuals neglect medical treatment due to the lack of specific symptoms. The lack of treatment may allow chlamydial organisms to ascend to the upper genital tract, causing pelvic inflammatory diseases, ectopic pregnancy, or tubal infertility, depending on the upper genital tissues affected and the severity of the invasion by the chlamydial organisms. The current clinical challenges include no effective means for preventing chlamydial ascension and no approved C. trachomatis vaccines for preventing chlamydial infection or disease. Modeling chlamydial infection in the female mouse genital tract has provided a productive platform for revealing pathogenic and protective mechanisms during chlamydial infection [1].
The mouse-adapted C. muridarum can efficiently evade innate immunity in the lower genital tract and ascend to the upper genital tract where it induces long-lasting sequelae [2,3] such as hydrosalpinx, which is similar to that observed under laparoscopy in women infected with the human pathogen C. trachomatis. Thus, the C. muridarum model has been used to investigate the pathogenic mechanisms of long-term chlamydia infection [4–8]. In contrast, infection of the female mouse genital tract with C. trachomatis is limited and often fails to induce oviduct pathology [9]. Nevertheless, intravaginal inoculation with a high dose of C. trachomatis or transcervical inoculation with a moderate dose can cause glandular duct dilation, a pathology that was initially identified in C57 mice infected with C. muridarum [10]. Glandular duct dilation is observed as uterine horn dilation macroscopically. Since hydrosalpinx and uterine horn dilation can both lead to infertility, these pathologies have been used for evaluating chlamydial pathogenicity. Therefore, in this study, we used mouse models to study the pathology of the endometrium and oviduct following C. muridarum inoculation.
Materials and reagents
Biological materials
1. Mice: female (15–18 g), ≥ 6 weeks old, with strains and genotypes (wild type, transgenic, or knockout) to be determined depending on the specific experiment and model chosen. Mice were bred and maintained under specific pathogen-free conditions and on a standard chow diet for 8 weeks starting at 6 weeks old in the institutional animal facility of Shanghai Institute of Immunity and Infection
2. HeLa cells (human cervical epithelial carcinoma cells) (ATCC, catalog number: CCL2)
Reagents
1. Medroxyprogesterone acetate (Depo-Provera, Amphastar Pharmaceuticals, Inc, catalog number: 5401); resuspend in sterile 1× PBS (see Recipes) at a final concentration of 25 mg/mL
2. MD-76®R (NDC, catalog number: 0019-1317-09)
3. Trypan blue staining solution (Servicebio, catalog number: G1019-10ML)
4. 0.25% Trypsin-EDTA (1×), (Gibco, catalog number: 25200056)
5. 4% paraformaldehyde (Beyotime, catalog number: P0099-500 ml)
6. 0.1% Triton-X (Biosharp, catalog number: BL934B)
7. 3% BSA (COOLABER SCIENCE&TECHNOLOGY, catalog number: SL1331)
8. Disposable vacuum filters (500 mL, 0.22 μm) (BIOFIL, FMC201500)
9. DMEM (Gibco, catalog number: 11965092)
10. Heat-inactivated fetal bovine serum (FBS) (ExCell, catalog number: FND500)
11. Gentamicin, 10 mg/mL (BBI, catalog number: A428430)
12. Cycloheximide, 10 mg/mL (MCE, catalog number: HY-12320)
13. Sucrose (Sigma-Aldrich, catalog number: S9378)
14. Potassium phosphate, monobasic (Sigma-Aldrich, catalog number: P5655)
15. Potassium phosphate, dibasic trihydrate (Sigma-Aldrich, catalog number: P9666)
16. L-glutamic acid monosodium salt (Sigma-Aldrich, catalog number: G1626)
17. Sodium phosphate, dibasic (BBI, catalog number: A610879)
18. Sodium chloride (BBI, catalog number: A610476)
19. Potassium chloride (BBI, catalog number: A610440)
20. Potassium phosphate, monobasic (BBI, catalog number: A424391)
Solutions
1. Cycloheximide + gentamicin (C&G) growth medium (see Recipes)
2. MD-76R solutions, 52%, 44%, 40% (see Recipes)
3. Sucrose-phosphate-glutamate (SPG) buffer (see Recipes)
4. 1× PBS (see Recipes)
Recipes
1. Cycloheximide + gentamicin (C&G) growth medium
| Reagent | Volume |
|---|---|
| DMEM | 500 mL |
| Heat-inactivated fetal bovine serum | 50 mL |
| Gentamicin, 10 mg/mL | 1 mL |
| Cycloheximide, 10 mg/mL | 250 μL |
Store at 4 °C.
2. MD-76R solutions, 52%, 44%, 40%
| Solution | Reagents |
|---|---|
| 52% MD-76R | 52 mL MD-76R + 48 mL ddH2O |
| 44% MD-76R | 44 mL MD-76R + 56 mL ddH2O |
| 40% MD-76R | 40 mL MD-76R + 60 mL ddH2O |
Adjust to a total volume of 100 mL with sterile water and store at 4 °C.
3. Sucrose-phosphate-glutamate (SPG) buffer
| Reagent | Quantity |
|---|---|
| Sucrose | 74.62 g |
| Potassium phosphate, monobasic | 0.51 g |
| Potassium phosphate, dibasic trihydrate | 1.23 g |
| L-glutamic acid monosodium salt | 0.82 g |
| ddH2O | 1,000 mL |
| Total | 1,000 mL |
Mix with a stir bar until homogenous. Then, filter-sterilize the SPG buffer using disposable vacuum filters (500 mL, 0.22 μm, BIOFIL, FMC201500) and store at room temperature.
4. 1× PBS, pH 7.2
| Reagent | Quantity |
|---|---|
| Sodium phosphate, dibasic | 1.44 g |
| Sodium chloride | 8 g |
| Potassium chloride | 0.2 g |
| Potassium phosphate, monobasic | 0.24 g |
| ddH2O | 1,000 mL |
| Total | 1,000 mL |
Mix with a stir bar until homogenous. Sterilize PBS buffer by autoclaving and store at room temperature.
Laboratory supplies
1. 1 mL syringe & 27-gauge needle (Becton, Dickinson and Company, catalog number: 305109)
2. 1.5 mL microcentrifuge tubes (e.g., Fisher Scientific, catalog number: 21-402-903 or equivalent)
3. Speculum and non-surgical embryo transfer device (NSET) (ParaTechs, catalog number: 60010 or equivalent)
4. 96-well flat bottom cell culture plates, TC-treated (BIOFIL, catalog number: TCP010096)
5. Cell counting plate (Countstar, catalog number: CO010101)
Equipment
1. Microscope (YueHe, model: YHF40)
2. Centrifuge for 96-well plate (Eppendorf, model: Centrifuge 5810 R)
3. Laminar flow hood (Nuaire BSC Class II or equivalent)
4. Multi-channel pipettor (RAININ, model: L12-200XLS+)
5. P20 micropipette (Gilson, model: TB27868 or equivalent)
6. Fluorescence microscope (Olympus, model: AX70) with a CCD camera (Hamamatsu)
7. Ultracentrifuge (Beckman Coulter, model: Optima L-100K) with swinging bucket rotor (Beckman, model: SW 28 Ti or equivalent)
8. Superspeed centrifuge (Thermo Scientific, model: Sorvall RC 6 Plus) with fixed-angle rotor (Sorvall SS 34 or equivalent)
9. Sonics VCX 130 sonicator (Sonics & Materials, model: VCX 130 or equivalent).
10. Countstar Altair (Shanghai Ruiyu Biotechnology Co., Ltd)
Procedure
A. Amplification and purification of chlamydial organisms
1. Chlamydial organisms are amplified in cell culture and purified as EBs using a gradient centrifugation method, as briefly described below:
a. Prepare a gradient from bottom to top in a tube with 5 mL of 52% MD-76R, 8 mL of 44% MD-76R, and 13 mL of 40% MD-76R (see Recipes).
b. Sonicate the infected cells 8–15 times at 60 W for 30 s each to release EBs from cells.
c. Centrifuge the sonicated lysates at 1,200× g for 10 min at 4 °C to remove the cell pellet.
d. Load 10 mL of the supernatant on top of each gradient using the same method as for laying MD-76R layers.
e. Ultracentrifuge at 17,000× g for 90 min at 4 °C in Beckman SW 28 Ti rotor (slow acceleration and brakes off).
f. The EB band can be seen as a cloudy white/yellow band in the 52%–44% interphase after ultracentrifuge.
g. Resuspend gradient-purified EBs in SPG buffer (see Recipes) and store in aliquots at -80 °C until use.
B. Intravaginal inoculation with C. muridarum EBs
1. Five days before inoculation with C. muridarum EBs, clean mouse abdomen with 70% (v/v) ethanol and inject 100 μL of 25 mg/mL medroxyprogesterone acetate with a 27- or 30-gauge needle by subcutaneous injection (I.s). Observe mouse for 2–3 min for any signs of adverse effects.
2. Take a C. muridarum stock aliquot from the -80 °C freezer and thaw on ice. Using sterile techniques, operate the following steps inside a laminar flow hood.
3. Dilute the stock with sterile SPG buffer to the desired 2 × 105 inclusion-forming units (IFUs) per 10 μL. Mix well and keep on ice until mouse inoculation.
Note: For example, if the stock titer is 1 × 107 IFU/µL, first dilute the stock in 1:10 (10 μL + 90 μL SPG buffer) for a stock titer of 1× 106 IFU/µl. Then, continue to dilute the stock in 1:50 (20 μL + 980 μL SPG buffer) for a final stock titer of 2 × 104 IFU/µL (2 × 105 IFUs per 10 μL).
4. Use a p20 micropipette to pipette 10 μL of solution from the tube that contains the desired C. muridarum EBs. Place the micro-pipettor flat on the right-hand side.
5. Use the left hand to hold the mouse’s belly up, and the head tilted downward (Figure 1A).
6. Use the right hand to gently insert the micro-pipettor tip (containing 10 μL of inoculum) into the vagina until a slight resistance is felt (from the cervix). Slowly eject the inoculum until the first stop is reached. Hold the micro-pipettor without releasing the plunger when removing the micropipette tip from the vagina (Figure 1B).
7. Continue to hold the mouse in the same position for 2 min to ensure inoculum remains in the vaginal vault (which may increase chlamydial invasion of vaginal and ectocervical epithelial cells).
8. After the 2 min, the mouse can be returned to the cage. A brief observation for any signs of adverse effects is useful for ensuring the quality of the procedure.
9. For evaluating pathology, euthanize the mice with CO2 for 2 min in the chamber and fix them on the dissection table. Using micro-scissors, incise the abdominal skin, abdominal muscle, and peritoneum along the midline.
10. Lift the intestinal tissue upward to expose the Y-shaped reproductive tract and surrounding adipose tissue. Use micro-scissors to cut the mouse's pubis and intermittently cut the lower section of the reproductive tract below the pubis (Figure 1C).
11. Then, gradually cut and lift the reproductive tract upward, separating it from the surrounding connective tissue.
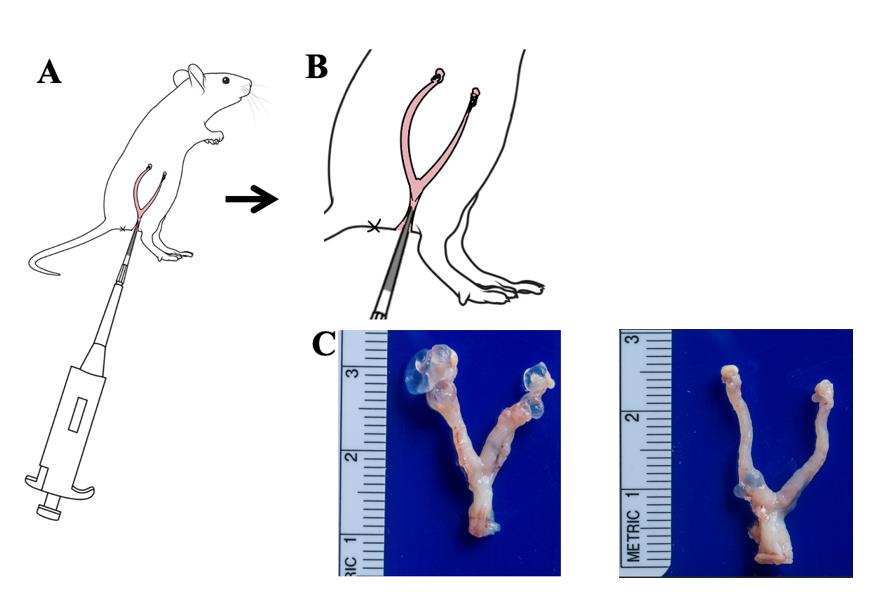
Figure 1. Key steps of intravaginal inoculation of mice genital tract with C. muridarum. A. Schematic diagram of intravaginal inoculation of C. muridarum in the mice genital tract. Mice were inoculated intravaginally with 2 × 105 IFUs of C. muridarum as described in section A. B. Schematic diagram of local magnification of genital tract infection manipulation in mice. C. Pathology after intravaginal inoculation; a group of mice was inoculated intravaginally with 2 × 105 IFUs of C. muridarum (n = 5, left) or SPG buffer (Ctrl, n = 5, right) as described in section A. On day 56, mice were sacrificed to observe upper genital tract pathology hydrosalpinx. Data from two or three independent experiments.
C. Transcervical inoculation with C. muridarum EBs
1. Five days before inoculation with C. muridarum EBs, clean the mouse’s abdomen with 70% (v/v) ethanol and inject 100 μL of 25 mg/mL medroxyprogesterone acetate intraperitoneally (i.p) with a 27- or 30-gauge needle. Observe the mouse for signs of adverse effects.
2. After five days, take a C. muridarum stock aliquot from the -80 freezer and thaw on ice. Using sterile techniques, operate the following steps inside a laminar flow hood.
3. Dilute stock with sterile SPG buffer to desired 2 × 105 IFUs per 10 μL. Mix well and keep on ice until mouse inoculation.
4. Connect the NSET device to the p20 micropipette and take up 10 μL of chlamydial EBs.
5. Gently place a speculum into the vagina to open up the mouse’s vagina. Insert the NSET device into the speculum and through the cervix. Press the pipette plunger completely to deliver chlamydial EB.
6. Remove the NSET device gently without releasing the pipette plunger and remove the speculum.
7. Monitor mice for several minutes for any adverse signs and then return them to the cage.
8. For evaluating cervix dilation, after euthanizing mice with CO2 in the chamber, fix mice on the dissection table. Using micro-scissors, incise the abdominal skin, abdominal muscle, and peritoneum along the midline.
9. Lift the intestinal tissue upward to expose the Y-shaped reproductive tract and surrounding adipose tissue. Use micro-scissors to cut the mouse's pubis and intermittently cut the lower section of the reproductive tract below the pubis.
10. Gradually cut and lift the reproductive tract upward, separating it from the surrounding connective tissue (Figure 2).
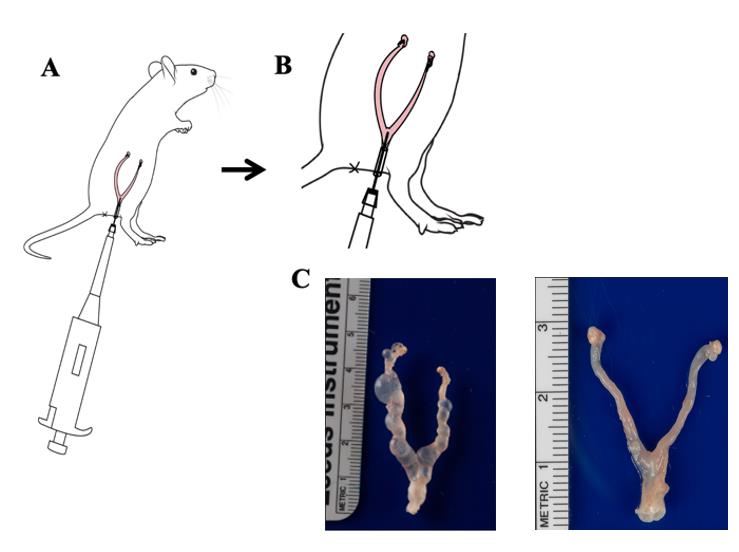
Figure 2. Key steps of transcervical inoculation of mice genital tract with C. muridarum. A. Schematic diagram of transcervical inoculation of C. muridarum in the mice genital tract. Mice were inoculated with 2 × 105 IFUs of C. muridarum as described in section B. B. Schematic diagram of local magnification of transcervical inoculation manipulation in mice. C. Pathology after transcervical inoculation; mice were inoculated with 2 × 105 IFUs of C. muridarum (n = 5, left) or SPG buffer (n = 5, right) as described in section B. On day 56, mice were sacrificed for observing uterine horn dilation. Data from two or three independent experiments.
D. HeLa cells preparation
1. Prepare a 175 cm flask seeded with 1-day-old HeLa cells at approximately 90% confluency.
2. Wash Hela cells once with 1× PBS and treat with 0.25% Trypsin-EDTA (1×) in a CO2 incubator for 2 min. Add 10 mL of DMEM +10% FBS medium to the 175 cm2 flask and mix the cells after Trypsin-EDTA treatment.
3. Set up a HeLa monolayer 96-well microplate with 2.5 × 104 cells/well. Make sure that monolayer confluency is kept between 70%–80% at the time of C. muridarum infection.
Note: 10 μL of cell suspension + 10 μL of trypan blue staining solution (total 20 μL) was automatically counted using a Countstar Altair.
E. Detecting the load of live chlamydial organisms
1. The day after setting up the HeLa monolayer microplate, add 500 μL of sterile SPG buffer and 3–4 autoclaved glass beads to a 1.5 mL microcentrifuge tube in the flow hood.
2. Label the tube with the ID of the mouse that will be swabbed.
3. Insert a sterile rayon mini-tipped applicator into the vaginal vault without pushing past the cervix. A slight resistance can be felt when the swab touches the cervix.
4. Rotate the swab clockwise and counterclockwise five times in each direction.
5. Pull the swab out and place it in the pre-labeled 1.5 mL microcentrifuge tube. Cut the handle of the applicator short enough to close the microcentrifuge cap and immediately put the tube on ice for subsequent processing.
6. Thoroughly vortex swab samples for 2 min at high speed to release chlamydial organisms into solution. Then, prepare a 96-well flat bottom TC-treated plate to be used for serially diluting the swab sample.
7. After serial dilution (from 1:10 to 1:80 by 2-fold serial dilutions), transfer 100 μL of the serially diluted swab samples onto the 96-well plate with 24-hours-old HeLa cell monolayer using a multi-channel pipettor starting from the highest diluted row.
8. Centrifuge the 96-well plates infected with chlamydial samples at 800× g for 1 h at room temperature.
9. Remove inoculum and add 200 μL of C&G growth medium to the 96-well plates.
10. Incubate the plates in a 37 °C, 5% CO2 tissue culture incubator . After 20–24 h of incubation, the microplates are ready for processing to visualize chlamydial inclusions using an immunofluorescence assay.
Note: Here, the primary antibody (rabbit antibody) was made with purified C. muridarum elementary bodies and was a gift from Jingyue Ma from Tianjin Medical University General Hospital. After primary antibody incubation, the secondary antibody (goat anti-rabbit IgG conjugated with Cy2, Abcam, #ab6940) was added, and nuclei were visualized using the DNA dye Hoechst 33258 (Abcam, #ab228550). The doubly labeled samples were used for counting chlamydial inclusions under a fluorescence microscope with a CCD camera.
11. Fix the infected HeLa cells with 100 μL of 4% paraformaldehyde and incubate for 40–60 min at room temperature.
12. Remove paraformaldehyde and wash the plate with 1× PBS once. Then, permeabilize the fixed cell monolayers with 100 μL of 0.1% Triton-X for 30 min at room temperature.
13. Block the permeabilized cells with 100 μL of 3% BSA for 1 h at 37 °C or overnight at 4 °C after removing the Triton-X solution.
14. Label chlamydial antigens with a rabbit polyclonal antibody raised with C. muridarum EBs as the primary antibody: add 50 μL of the rabbit antibody and incubate at 37 °C for 1 h or overnight at 4 °C.
15. Remove the primary antibody solution and wash the plate with 1× PBS three times.
16. Detect the immobilized primary antibody by adding 50 μL of the secondary antibody conjugate (goat anti-rabbit conjugated with FITC) solution that contains Hoechst DNA dye and incubate at 37 °C for 1 h.
17. Remove the secondary antibody solution and wash the plate with 1× PBS three times.
18. Calculate the total number of IFUs per swab or tissue based on the number of inclusions per well, dilution factor, and inoculation volumes (see sects E12–16).
F. Live chlamydial stock titration
1. One day prior to titration, seed cells at 1×105 cells per well in a 24-well tissue culture plate at approximately 90% confluency.
2. Make sure that monolayer confluency is kept between 70%–80% at the time of live chlamydial stock titration.
3. Prepare a 1:100 stock dilution by adding 10μL of EB stock to 990μL of SPG buffer. The starting 1:10,000 dilution was prepared by adding 10μL of 1:100 stock dilution to 990μL of SPG buffer. A 10-fold serial dilution was set up for as many serial dilutions as required (Figure 3A).
4. After serial dilution (from 104 to 109, by 10-fold serial dilutions), transfer 200 μL of the serially diluted samples onto the 24-well plate with 24-hour-old HeLa cell monolayer (Figure 3B).
5. Centrifuge the 24-well plates infected with chlamydial samples at 800× g for 1 h at room temperature.
6. Remove inoculum and add 200 μL of C&G growth medium to the 24-well plates.
10. Incubate the plates in a 37 °C, 5% CO2 tissue culture incubator for 20–24 h.
11. The microplates are ready for processing to visualize chlamydial inclusions using an immunofluorescence assay (see steps D10–17) (Figure 3C, D).
12. Count inclusions under a fluorescence microscope as follows.
13. Starting with the highest dilution at 10× magnification, count the number of inclusions in the center, upper-right, lower-right, lower-left, and upper-left sections of the well.
14. If fewer than 10 inclusions are visible per field at 10× magnification, count the inclusions present in three wells. Conversely, if more than 25 inclusions are visible, use the next highest magnification and count inclusions.
15. Evaluate each dilution at the lowest magnification, increasing the magnification accordingly. Count five random views and record the dilution and magnification for each count.
16. Determine IFU/mL as described below:
IFU/mL=(IFU counted/Fields counted) (Dilution)[ πr2/π(A/2)2]/Inoculation vol (mL)−1
Where: r2=(well radius)2
FOV=field of vision
A=FOV eye piece/objective magnification
π(A/2)2=Area of radius viewed
Note 1. When sample infection rate reaches 100%, these samples are not included in the count.
Note 2. The cell monolayer was not used to count the number of inclusions (does not cover the entire field or has swaths missing).
Note 3. If your sample is diluted appropriately, an inclusion is the result of an EB infection of a HeLa cell, which is designated as an inclusion-forming unit or IFU.
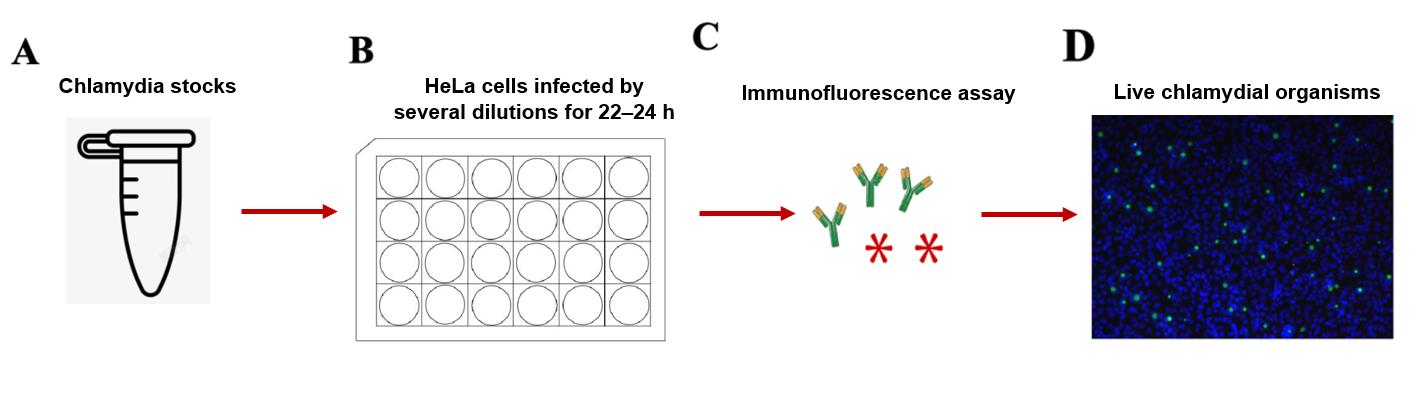
Figure 3. Live chlamydial titration by immunofluorescence assay. A. Prepare the chlamydia stock. B. HeLa cells are infected by several dilutions for 22–24 h. C. The cell plate is set up by immunofluorescence assay. D. The fluorescence signal is detected under a fluorescence microscope.
Data analysis
The evaluation of hydrosalpinx in mice following intravaginal inoculation is performed as follows (Table 1).
Table 1. Hydrosalpinx score
| Score | Pathology |
|---|---|
| 0 | No hydrosalpinx |
| 1 | Hydrosalpinx only observable under magnification |
| 2 | Visible hydrosalpinx smaller than the size of ovary |
| 3 | Visible hydrosalpinx roughly equal the size of ovary |
| 4 | Visible hydrosalpinx larger than the size of ovary |
Validation of protocol
This protocol was validated by the Guangming Zhong lab, and the resulting work was published in (Figures 4, 5):
Sun et al. [10]. Chlamydia muridarum induction of glandular duct dilatation in mice. Infect Immun.
Chen et al. [3]. Chlamydial induction of hydrosalpinx in 11 strains of mice reveals multiple host mechanisms for preventing upper genital tract pathology. PLoS One.
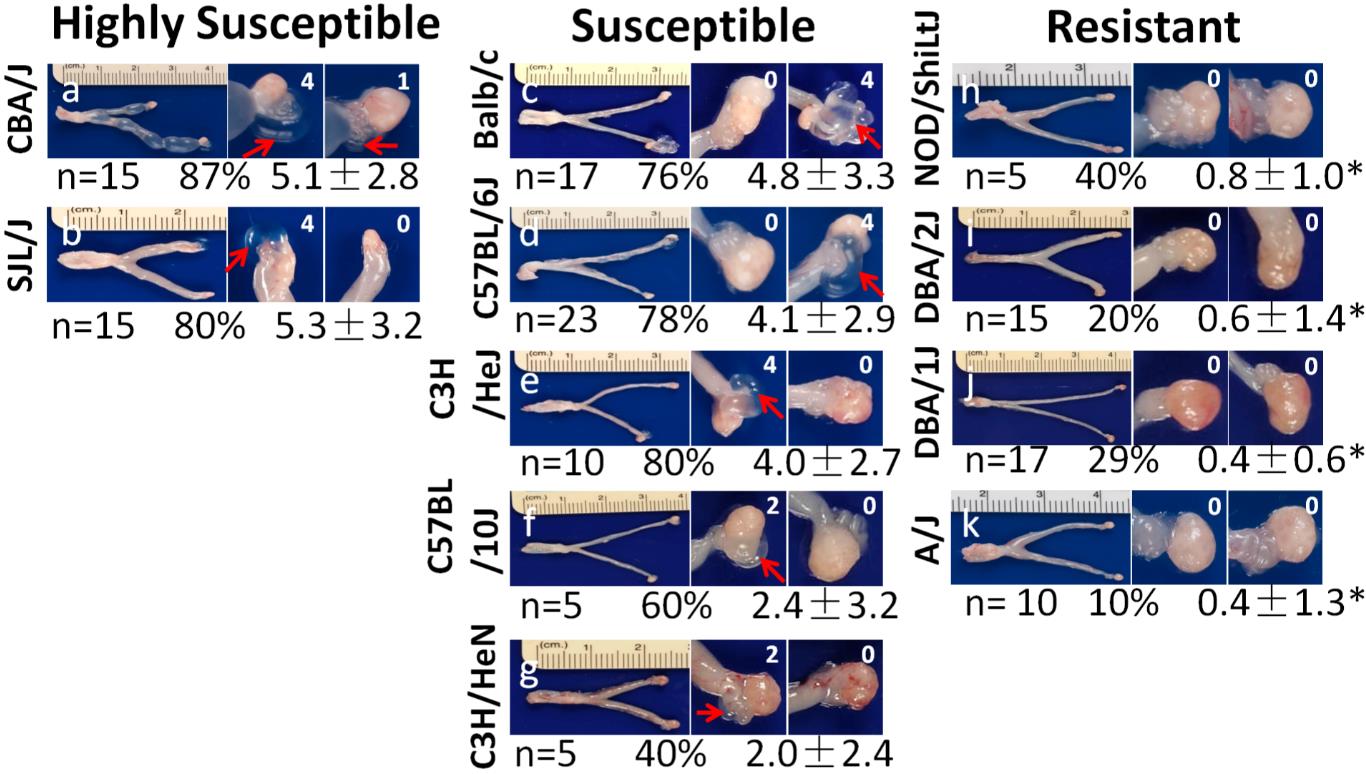
Figure 4. Data example showing hydrosalpinx development in 11 strains of mice following lower genital tract infection with C. muridarum. Representative image from each strain of mice is presented with the whole genital tract in the left and the magnified oviduct/ovary portion in the right. The number of mice with positive hydrosalpinx (as marked with red arrows in the magnified oviduct/ovary images) were counted and recorded as a percentage of hydrosalpinx-positive mice. Reprinted/adapted from Chen et al. [3]. 2014 The Authors. Published by PLoS One.
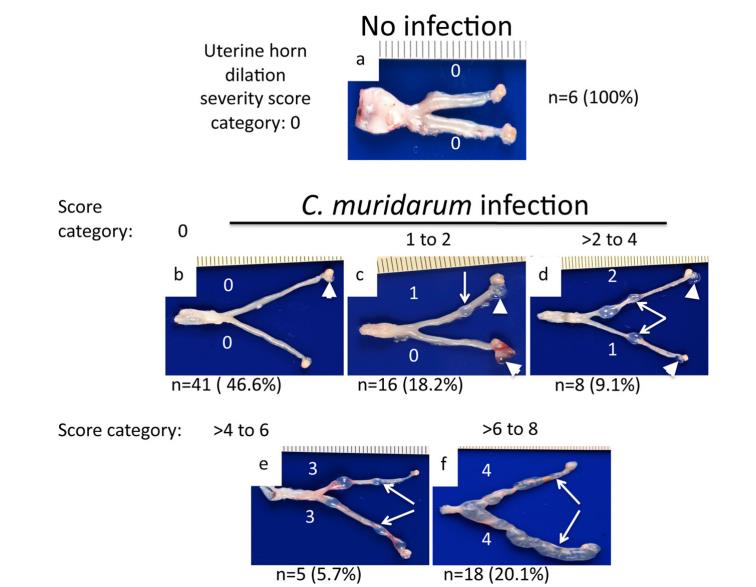
Figure 5. Data example showing C. muridarum induction of uterine horn dilation. Reprinted/adapted from Sun et al. [10]. 2015 The Authors. Published by Infection and Immunity.
General notes and troubleshooting
1. For the gradient purification of EBs, all solutions and centrifuge tubes must be pre-chilled at 4 °C or on ice prior to usage; they should remain on ice during usage. Rotors and buckets utilized during centrifugation must be cleaned with 70% ethanol and pre-chilled prior to use.
2. All steps must be performed aseptically in biological safety cabinets to avoid contamination.
3. For the intravaginal inoculation, suggested inoculation concentrations are 2 × 105 for C. muridarum. It is recommended to titrate your stock in mice before formal experiments.
4. Ensure the micropipette tip is not inserted past the cervix during intravaginal inoculation as this can cause unnecessary pain and injury to the mouse.
5. For the transcervical inoculation, follow the manufacturer’s instructions when using the NSET device.
Acknowledgments
We would like to express our heartfelt thanks to Prof. Guangming Zhong for his invaluable guidance and support throughout our research. We also wish to acknowledge the contributions of his lab members, particularly Jianlin Chen, Hongbo Zhang, Zhou Zhou, and Zhangsheng Yang, for their assistance and collaboration in improving the techniques used in this study. Their expertise and dedication have been instrumental in the success of our work. This work was supported by the National Natural Science Foundation of China for the Youth (32100162) to Q.T. and (32000138) to T.Z.
Competing interests
We declare no conflict of interest or competing interests.
Ethical considerations
All animal studies have been approved by the Ethics Committee of the Institute of Immunity and Infection of Shanghai Chinese Academy of Sciences (A2020019).
References
- Wang, Y., He, R., Winner, H., Gauduin, M. C., Zhang, N., He, C. and Zhong, G. (2023). Induction of Transmucosal Protection by Oral Vaccination with an Attenuated Chlamydia. Infect Immun. 91(5): e00043–23. https://doi.org/10.1128/iai.00043-23
- Campbell, J., Huang, Y., Liu, Y., Schenken, R., Arulanandam, B. and Zhong, G. (2014). Bioluminescence Imaging of Chlamydia muridarum Ascending Infection in Mice. PLoS One. 9(7): e101634. https://doi.org/10.1371/journal.pone.0101634
- Chen, J., Zhang, H., Zhou, Z., Yang, Z., Ding, Y., Zhou, Z., Zhong, E., Arulanandam, B., Baseman, J., Zhong, G., et al. (2014). Chlamydial Induction of Hydrosalpinx in 11 Strains of Mice Reveals Multiple Host Mechanisms for Preventing Upper Genital Tract Pathology. PLoS One. 9(4): e95076. https://doi.org/10.1371/journal.pone.0095076
- Tian, Q., Zhou, Z., Wang, L., Abu-Khdeir, A. M., Huo, Z., Sun, X., Zhang, N., Schenken, R., Wang, Y., Xue, M., et al. (2020). Gastrointestinal Coinfection Promotes Chlamydial Pathogenicity in the Genital Tract. Infect Immun. 88(4): e00905–19. https://doi.org/10.1128/iai.00905-19
- Conrad, T. A., Gong, S., Yang, Z., Matulich, P., Keck, J., Beltrami, N., Chen, C., Zhou, Z., Dai, J., Zhong, G., et al. (2016). The Chromosome-Encoded Hypothetical Protein TC0668 Is an Upper Genital Tract Pathogenicity Factor of Chlamydia muridarum. Infect Immun. 84(2): 467–479. https://doi.org/10.1128/iai.01171-15
- Yang, C., Lei, L., Collins, J. W. M., Briones, M., Ma, L., Sturdevant, G. L., Su, H., Kashyap, A. K., Dorward, D., Bock, K. W., et al. (2021). Chlamydia evasion of neutrophil host defense results in NLRP3 dependent myeloid-mediated sterile inflammation through the purinergic P2X7 receptor. Nat Commun. 12(1): 5454. https://doi.org/10.1038/s41467-021-25749-3
- Yang, C., Kari, L., Lei, L., Carlson, J. H., Ma, L., Couch, C. E., Whitmire, W. M., Bock, K., Moore, I., Bonner, C., et al. (2020). Chlamydia trachomatis Plasmid Gene Protein 3 Is Essential for the Establishment of Persistent Infection and Associated Immunopathology. mBio. 11(4): e01902–20. https://doi.org/10.1128/mbio.01902-20
- Dockterman, J., Reitano, J. R., Everitt, J. I., Wallace, G. D., Hendrix, M., Taylor, G. A. and Coers, J. (2024). Irgm proteins attenuate inflammatory disease in mouse models of genital Chlamydia infection. mBio. 15(4): e00303–24. https://doi.org/10.1128/mbio.00303-24
- Sturdevant, G. L. and Caldwell, H. D. (2014). Innate immunity is sufficient for the clearance ofChlamydia trachomatisfrom the female mouse genital tract. Pathog Dis. 72(1): 70–73. https://doi.org/10.1111/2049-632x.12164
- Sun, X., Yang, Z., Zhang, H., Dai, J., Chen, J., Tang, L., Rippentrop, S., Xue, M., Zhong, G., Wu, G., et al. (2015). Chlamydia muridarum Induction of Glandular Duct Dilation in Mice. Infect Immun. 83(6): 2327–2337. https://doi.org/10.1128/iai.00154-15
Article Information
Publication history
Received: Aug 3, 2024
Accepted: Nov 28, 2024
Available online: Dec 26, 2024
Published: Feb 5, 2025
Copyright
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Wang, Y., Han, Z., Wang, L., Sun, X., Tian, Q. and Zhang, T. (2025). Development and Validation of Chlamydia muridarum Mouse Models for Studying Genital Tract Infection Pathogenesis. Bio-protocol 15(3): e5181. DOI: 10.21769/BioProtoc.5181.
Category
Microbiology > in vivo model > Protozoan
Cell Biology > Model organism culture
Medicine > Inflammation
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









