- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Identification of Mycobacterium tuberculosis and its Drug Resistance by Targeted Nanopore Sequencing Technology
Published: Vol 15, Iss 3, Feb 5, 2025 DOI: 10.21769/BioProtoc.5170 Views: 2025
Reviewed by: Alka MehraSoumya MoonjelySuresh Panthee

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Human, Bacterial and Fungal Amplicon Collection and Processing for Sequencing
Julia Oh
May 20, 2015 13700 Views

ChIP-seq Experiment and Data Analysis in the Cyanobacterium Synechocystis sp. PCC 6803
Joaquín Giner-Lamia [...] Matthias E. Futschik
Jun 20, 2018 13294 Views

Strand-specific Single-stranded DNA Sequencing (4S-seq) of E. coli genomes
Takahiro Masuda [...] Kazuharu Arakawa
Aug 5, 2019 7524 Views
Abstract
Tuberculosis (TB) remains the leading cause of human mortality in infectious diseases. Drug-resistant TB, particularly multidrug-resistant TB and extensively drug-resistant TB, poses a pressing clinical and public health challenge. The main causative agents of TB are known as Mycobacterium tuberculosis (MTB), which exhibits a highly complex drug resistance profile. Traditional culture-based phenotypic drug susceptibility testing is time-consuming, and PCR-based assays are restricted to detecting known mutational hotspots. In this study, we present a protocol leveraging high-throughput nanopore sequencing technology in conjunction with multiplex PCR, termed targeted nanopore sequencing, for the identification of MTB and analysis of its drug resistance. Our method for MTB drug resistance assessment offers the benefits of being culture-free, efficient, high-throughput, and highly accurate, which could significantly aid in clinical patient management and the control of TB infections.
Key features
• Targeted nanopore sequencing detects 18 genes simultaneously linked to antibiotic resistance in MTB.
• The method provides broad drug resistance profiles for 14 first- and second-line anti-TB drugs without bacterial culture.
• The expedited turnaround time of the process is approximately 7.5 h with a detection limit of 102 bacteria/mL.
Keywords: Mycobacterium tuberculosisGraphical overview
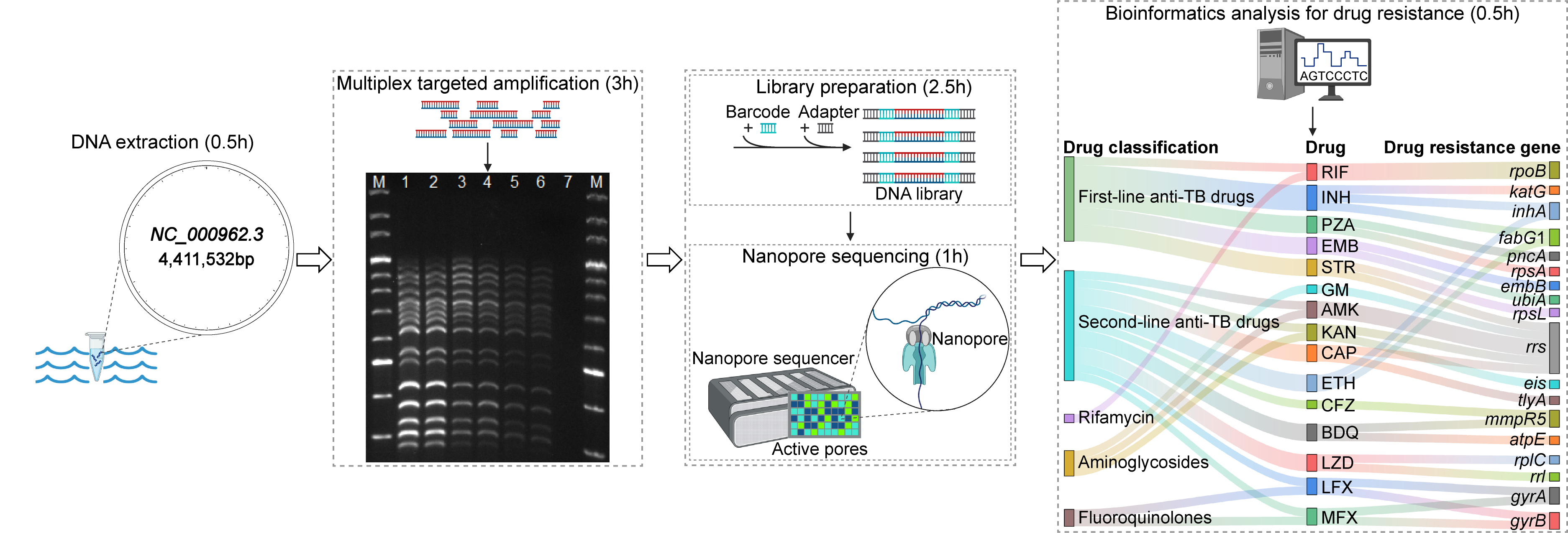
Targeted nanopore sequencing for the drug resistance assay of Mycobacterium tuberculosis. Initially, DNA is extracted, and the targeted regions within genes associated with drug resistance are amplified via multiplex PCR. Subsequently, the amplicons undergo barcoding and adapter ligation. The library preparation is then sequenced on a nanopore sequencer, which yields long-read sequence data. Ultimately, drug resistance is identified through bioinformatics analysis. The entire targeted nanopore sequencing process has an expedited turnaround time of approximately 7.5 h.
Background
Tuberculosis (TB) affirms its position as the preeminent cause of human death from infectious diseases, with an estimated 1.7 million deaths occurring globally each year, as reported by Daley [1]. The latest figures from the World Health Organization’s Global Tuberculosis Report 2023 reveal that 7.5 million new cases were diagnosed worldwide in 2022, surpassing pre-COVID levels. TB pathogen refers to a group of bacteria within the Mycobacterium tuberculosis complex (MTBC), including Mycobacterium tuberculosis (MTB), noted for its intricate drug resistance spectrum [2]. MTB, being the principal causative agent of TB, gains antibiotic resistance through gene mutations, significantly influencing clinical treatment approaches [3–5].
Surveillance of highly virulent and multidrug-resistant (MDR) MTB strains is imperative for securing precise diagnoses and potent treatment regimens [6]. Conventional culture-based phenotypic drug susceptibility testing (pDST) is the standard method for assessing MTB, although being time-consuming, typically requiring a span of days to weeks [3]. The prevalent molecular detection methods, including the real-time PCR-based Xpert MTB/RIF assay endorsed by the WHO, while being efficient, are limited to detecting known mutational hotspots and thus provide incomplete data on drug resistance [7]. Consequently, infections with MDR and extensively drug-resistant (XDR) MTB strains highlight an urgent need for advanced diagnostic tools to guide TB treatment options [8].
High-throughput sequencing technology has become a staple in clinical diagnostics. Nanopore sequencing, a cutting-edge method exemplified by the equipment from Oxford Nanopore Technologies (ONT), offers the benefits of long-read lengths, real-time data acquisition, and rapid sequencing [9]. While its accuracy has been a subject of improvement, significant strides in scientific and technological advancements have enhanced its performance in recent years [10,11]. Nanopore sequencing facilitates direct analysis of DNA or RNA sequences without the need for culturing MTB, enabling swift identification of MTB and detection of TB drug resistance. This approach might not only reduce the overall cost of MTB detection but also expedite the diagnostic process [12]. Therefore, the technology holds considerable promise for clinical applications and possesses a promising market outlook, making it an attractive option for clinical settings [13].
This study introduces a method that integrates multiplex PCR for targeted enrichment with nanopore sequencing, termed targeted nanopore sequencing. Furthermore, the bioinformatic tool TBProfiler is employed for drug resistance analysis, with its database encompassing 1,195 candidate mutations, which were obtained globally from more than 17,000 clinical isolates and assessed with whole-genome sequencing (WGS) and pDST [14].
Materials and reagents
Reagents
1. 10× TBE buffer (Solarbio, catalog number: T1051)
2. 1 M Tris-HCl (pH 8.0) (Solarbio, catalog number: T1150)
3. 200 bp DNA ladder (Dye Plus) (Takara, catalog number: 3423A)
4. Agarose (Mei5 Biotechnology, Co., Ltd, catalog number: MF103)
5. Agencourt AMPure XP beads (Beckman Coulter, catalog number: A63881)
6. Chelex 100 sodium (Solarbio, catalog number: C8230)
7. DNase/RNase-free water (Solarbio, catalog number: R1600)
8. dNTP mixture (Takara, catalog number: 4030)
9. EDTA (Sigma, catalog number: 798681)
10. Ligation Sequencing kit (Q20+) (Oxford Nanopore Technologies, catalog number: SQK-LSK114)
11. M5 6× DNA electrophoresis loading buffer (Mei5 Biotechnology, Co., Ltd, catalog number: MF144-01)
12. M5 Hipure Next III Gelred (Mei5 Biotechnology, Co., Ltd, catalog number: MF380-01)
13. NaOH (Sigma, catalog number: 221465)
14. Native Barcoding kit 24 (Q20+) (Oxford Nanopore Technologies, catalog number: SQK-NBD114.24)
15. NEB Blunt/TA ligase master mix (New England Biolabs, catalog number: M0367L)
16. NEB Q5 Hot Start High-Fidelity DNA Polymerase (New England Biolabs, catalog number: M0493L)
17. NEBNext Ultra II end repair/dA-tailing module (New England Biolabs, catalog number: E7546S)
18. NEBNext quick ligation module (New England Biolabs, catalog number: E6056S)
19. Nonidet P40 substitute (NP-40) (Solarbio, catalog number: N8030)
20. PBS (Sigma, catalog number: P3813)
21. Qubit dsDNA HS Assay kit (Thermo Fisher Scientific, catalog number: Q32854)
22. Triton X-100 (Solarbio, catalog number: T8200)
Laboratory supplies
1. 0.2 mL PCR tubes (ideally in strips of 8) (Thermo Fisher Scientific, catalog number: AB0490)
2. 1.5 mL tubes (Thermo Fisher Scientific, catalog number: S348903)
3. Pipette tips (Thermo Fisher Scientific, catalog numbers: 02-707-426, 02-707-403, 02-707-438)
4. PromethION flow cell R10.4.1 (Oxford Nanopore Technologies, catalog number: FLO-PRO114M)
5. Qubit assay tubes (Thermo Fisher Scientific, catalog number: Q32856)
Equipment
1. Biological safety cabinet (NuAire Lab Equipment, catalog number: NU-425-300)
2. Electrophoresis unit of power supply (Thermo Fisher Scientific, catalog number: S65533Q)
3. Magnetic stand for 1.5 mL tubes (Beckman Coulter, catalog number: A29182)
4. Microcentrifuge (Eppendorf, catalog number: EP5401000137)
5. Milli-Q ultrapure water system (Millipore Synergy, catalog number: F1CA45528 A)
6. NanoDrop spectrophotometer (Thermo Fisher Scientific, catalog number: ND-1000)
7. Qubit 4 fluorometer (Thermo Fisher Scientific, catalog number: Q33238)
8. Sequencing device (Oxford Nanopore Technologies, model: PromethION)
9. S1000 thermal cycler (Bio-Rad, catalog number: 1852196)
10. Vortex mixer (Thermo Fisher Scientific, catalog number: S96461A)
Software and datasets
1. Albacore (https://github.com/Albacore/albacore)
2. Bowtie2 (https://github.com/BenLangmead/bowtie2)
3. Delly (https://github.com/dellytools/delly)
4. MinKNOW (https://github.com/nanoporetech/minknow_api)
5. NanoFilt (https://github.com/wdecoster/nanofilt)
6. SAMtools (https://github.com/samtools/samtools)
7. TBProfiler (https://github.com/jodyphelan/TBProfiler)
8. Trimmomatic (https://github.com/usadellab/Trimmomatic)
Procedure
A. DNA extraction
1. Pre-treat clinical samples such as sputum and bronchoalveolar lavage fluid with 4% NaOH and PBS solution.
2. Collect 1 mL of the sample liquid into a 1.5 mL tube and centrifuge it at 11,000× g for 5 min; then, remove 950 μL of the supernatant.
3. Add 50 μL of nucleic acid extraction reagent (1 mM EDTA, 10 mM Tris-HCl, 1% NP-40, 1% Triton X-100, and 50% Chelex 100) to the tube and vortex the mixture for 1 min. The role of Chelex 100 is to capture divalent cations from biological samples and inhibit the activity of metallonucleases, even in highly concentrated salt solutions.
4. Place the mixture tube in a metal bath and boil it at 100 °C for 10 min.
5. After boiling, vortex the tube for 1 min and then centrifuge it at 11,000× g for 5 min.
6. Take the supernatant, enriched in DNA, as the template for multiplex PCR.
B. Multiplex PCR
1. Prepare a primer mixture containing primers designed for 18 drug resistance–associated genes, namely gyrB, gyrA, rpoB, mmpR5, rpsL, rplC, atpE, rrs, rrl, fabG1, inhA, rpsA, tlyA, katG, pncA, eis, embB, and ubiA, with their primer sequences and concentrations described in our previous publication [15].
2. Combine 1 μL of the multi-primer mixture, 0.25 μL of Q5 Hot-Start High-Fidelity DNA Polymerase, 5 μL of High GC enhancer, 5 μL of reaction buffer, 2 μL of dNTPs (2.5 mM), 4 μL of DNase/RNase-free water, and 10 μL of DNA template. Mix by flicking and spin down.
3. Perform the PCR with an initial denaturation step at 98 °C for 2 min, followed by 40 cycles of denaturation at 95 °C for 25 s, annealing at 60 °C for 30 s, and extension at 72 °C for 3 min, concluding with a final extension step at 72 °C for 4 min.
4. The amplicons can be visualized by agarose gel electrophoresis and quantified using a Qubit assay.
C. Library preparation
1. Purification of PCR amplicons
a. Take the AMPure XP beads out of the refrigerator at 4 °C, vortex to mix them evenly, and allow them to equilibrate at room temperature (RT) for at least 30 min before use.
b. Adjust the volume of 24 PCR amplicon samples to the lowest concentration observed, then add equal volumes to 24 × 1.5 mL tubes, supplementing with nuclease-free water to a final volume of 24 μL per tube.
c. To each of the 24 tubes, add 36 μL (1.5×) of the AMPure XP beads for DNA purification purposes, mix by pipetting 10 times, and incubate at RT for 15 min.
d. Place the tubes on a magnetic stand for 2–3 min, then carefully remove and discard the supernatant, ensuring the beads are not disturbed.
e. Gently add 200 μL of freshly prepared 80% ethanol to each tube, incubate at RT for 30–60 s, then remove and discard the ethanol. Air dry the beads for 2–3 min and repeat the process twice.
f. Remove the tubes from the magnetic stand, resuspend beads in 24 μL of nuclease-free water, and mix gently by flicking or pipetting up and down five times. Incubate for 5 min at RT.
g. Place the tubes back on the magnetic stand and carefully transfer 22.4 μL of the supernatant containing the eluted DNA to new tubes after the solution clears.
h. Perform quantification using a Qubit assay.
2. DNA end repair
a. Add 1.4 μL of Ultra II end-prep reaction buffer and 1.2 μL of Ultra II end-prep enzyme mix (from NEBNext end repair module) to the tubes containing supernatant with eluted DNA to achieve repaired DNA with 5' phosphorylated, 3' dA-tailed ends.
b. Gently mix the solution by pipetting up and down five times and incubate at 20 °C for 50 min, followed by a 20-min incubation at 65 °C in a thermocycler.
c. Follow the same bead addition and incubation steps as in steps C1c, d, and e.
d. Resuspend the beads in 16 μL of nuclease-free water, mix gently, and incubate for 5 min at RT.
e. Place the tubes back on the magnetic stand and carefully transfer 14.5 μL of the supernatant with eluted DNA to new tubes after the solution clears.
f. Perform quantification using a Qubit assay.
3. Ligation of barcodes
a. Add 1.5 μL of barcodes (Native Barcoding kit 24) and 15 μL of Blunt/TA ligase (NEB Blunt/TA ligase master mix) to the tubes with supernatant containing eluted DNA, providing 24 unique barcodes to enable multiplexing of amplicons.
b. Gently mix the solution and incubate at 20 °C for 60 min in a thermocycler, mixing every 20 min.
c. Follow the same bead addition and incubation steps as in steps C1c, d, and e.
d. Resuspend the beads in 10 μL of nuclease-free water, mix gently, and incubate for 5 min at RT.
e. Place the tubes back on the magnetic stand and carefully transfer 10 μL of the supernatant with eluted DNA to new tubes after the solution clears.
f. Perform quantification using a Qubit assay.
4. Ligation of sequencing adapters
a. Combine 24 PCR products into a new tube to create a library pool with an equal amount of DNA.
b. To the 25 μL library pool, add 10 μL of ligation buffer, 5 μL of NEBNext Quick T4 DNA ligase, and 5 μL of native adapter from the Oxford Nanopore Ligation Sequencing kit. Mix gently and briefly spin down the reaction.
c. Incubate the library pool for 60 min at 20 °C in a thermocycler.
d. Follow the same bead addition and incubation steps as in steps C1c, d, and e.
e. Add 200 μL of short fragment buffer to the library pool, incubate for 1 min, and carefully discard the supernatant. Repeat the process twice.
f. Resuspend the beads in 22 μL of elution buffer provided in the Ligation Sequencing kit and mix gently.
g. Spin down the pool and incubate for 10 min at RT.
h. Place the pool back onto the magnetic stand for 1 min and then transfer the eluate with the DNA library to a new tube, discarding the beads.
i. Perform quantification using a Qubit assay; the concentration is approximately 10 ng/μL.
D. Nanopore sequencing
1. Combine 30 μL of flow cell tether (Ligation Sequencing kit) with 1,170 μL of flow cell flush to prepare the priming mix, which is used for initiating flow cells with enough active pores for sequencing.
2. Apply 600 μL of the priming mix to the PromethION flow cell (R10.4.1) with R10 nanopores for high-consensus accuracy sequencing, wait for 5 min, and then add the rest of the mix.
3. Mix 21 μL of the library, 34 μL of loading beads, and 50 μL of sequencing buffer by gently inverting the tube 10 times.
4. Load 105 μL of the prepared library into the PromethION flow cell (R10.4.1) and initiate DNA sequencing for 1 h as per the manufacturer's protocol.
E. Bioinformatics analysis
1. Primary data acquisition is performed using MinKNOW software, which operates nanopore sequencing devices with a feature-rich user interface and saves data in FAST5 file format.
2. Albacore conducts base recognition to convert data from FAST5 to raw sequence data in FASTQ file format.
3. Reads with a Phred quality value ≤ 9 are filtered out by NanoFilt.
4. The bioinformatics pipeline TBProfiler detects antibiotic resistance mutations and can be accessed from its GitHub repository (https://github.com/jodyphelan/TBProfiler), installed via Bioconda, and utilized for resistance analysis. In Phelan et al. [14], a schematic was provided, highlighting the main steps in the TBProfiler pipeline, as well as an example of the TBProfiler report, illustrating the final output.
a. Compile sequencing read datasets from public MTB genomes with known resistance profiles for both first- and second-line antibiotics.
b. Update the TBProfiler mutation library as needed.
c. In standard operation, reads are trimmed with Trimmomatic (parameters: LEADING:3, TRAILING:3, SLIDINGWINDOW:4:20, MINLEN:36) and aligned to the H37Rv reference genome (GenBank accession no. NC_000962.3) using Bowtie2 with default parameters.
d. Call variants with BCFtools mpileup (parameters: -ABq0, -Q0, -a DP, AD) and BCFtools call (parameters: -mg 10), annotate with BCFtools csq (parameters: -p m), and parallelize the process with GNU parallel. Deletion calling is performed using Delly.
e. TBProfiler generates reports in JSON, TXT, and PDF formats, organizes data for visualization on a phylogenetic tree with iTOL, and allows for the generation and upload of config files to iTOL for visualizing drug resistance types, lineage, and individual drug resistance predictions.
Validation of protocol
Targeted nanopore sequencing, an advanced method that integrates nanopore sequencing technology with multiplex PCR, is capable of sequencing 18 genes simultaneously linked to antibiotic resistance in MTB in approximately 7.5 h. This method not only differentiates MTB from other bacterial species but also achieves a detection limit of 102 bacteria/mL by analyzing the datasets of all 18 targeted genes from 102 bacteria/mL to 107 bacteria/mL. Additionally, it furnishes detailed drug resistance profiles for 14 first- and second-line anti-TB drugs, facilitating informed drug prescription (see Graphical overview). While PromethION was utilized in this context, alternative devices like the portable MinION could be employed. Comprehensive data analysis and discussion are available in the publication by Tang et al. [15], accessible through the provided DOI: https://doi.org/10.3389/fmicb.2024.1331656.
General notes and troubleshooting
1. In this protocol, a single-tube multiplex PCR is developed for simultaneous amplification of 18 genes, with the set of PCR primers screened and validated.
2. The read depth of each gene cannot be consistent across the 18 targeted genes, and by optimizing the sequence and the concentration of each primer pair in the primer mixture, the output data can achieve better performance.
Acknowledgments
The authors thank the Scientific Research Center of Wenzhou Medical University for consultation and instrument availability, the Department of Clinical Laboratory from Wenzhou Central Hospital for providing clinical samples, and the original research paper [15] in which this protocol was described and validated. This research was supported by the Zhejiang Provincial Natural Science Foundation of China (Grant No. LTGG24H200001) and the Research Initiation Fund of Wenzhou Medical University (Grant No. GTJ21023).
Competing interests
The authors declare that they have no conflicts of interest with the contents of this article.
Ethical considerations
The study design was approved by the Research Ethics Board of Wenzhou Central Hospital, and all analyses were performed following the Declaration of Helsinki (L2023-02-033).
References
- Daley, C. L. (2019). The Global Fight Against Tuberculosis. Thorac Surg Clin. 29(1): 19–25.
- Gagneux, S. (2018). Ecology and evolution of Mycobacterium tuberculosis. Nat Rev Microbiol. 16(4): 202–213.
- Koch, A., Cox, H. and Mizrahi, V. (2018). Drug-resistant tuberculosis: challenges and opportunities for diagnosis and treatment. Curr Opin Pharmacol. 42: 7–15.
- Singh, V. and Chibale, K. (2021). Strategies to Combat Multi-Drug Resistance in Tuberculosis. Acc Chem Res. 54(10): 2361–2376.
- Salari, N., Kanjoori, A. H., Hosseinian-Far, A., Hasheminezhad, R., Mansouri, K. and Mohammadi, M. (2023). Global prevalence of drug-resistant tuberculosis: a systematic review and meta-analysis. Infect Dis Poverty. 12(1): 57.
- Consortium, C., Allix-Beguec, C., Arandjelovic, I., Bi, L., Beckert, P., Bonnet, M., Bradley, P., Cabibbe, A. M., Cancino-Munoz, I., Caulfield, M. J., et al. (2018). Prediction of susceptibility to first-line tuberculosis drugs by DNA sequencing. N Engl J Med. 379(15): 1403–1415.
- Weinrick, B. (2020). Genotyping of Mycobacterium tuberculosis Rifampin Resistance-Associated Mutations by Use of Data from Xpert MTB/RIF Ultra Enables Large-Scale Tuberculosis Molecular Epidemiology Studies. J Clin Microbiol. 58(1): e01504–19.
- Wu, X., Liang, R., Xiao, Y., Liu, H., Zhang, Y., Jiang, Y., Liu, M., Tang, J., Wang, W., Li, W., et al. (2023). Application of targeted next generation sequencing technology in the diagnosis of Mycobacterium Tuberculosis and first line drugs resistance directly from cell-free DNA of bronchoalveolar lavage fluid. J Infect. 86(4): 399–401.
- Shendure, J., Balasubramanian, S., Church, G. M., Gilbert, W., Rogers, J., Schloss, J. A. and Waterston, R. H. (2017). DNA sequencing at 40: past, present and future. Nature. 550(7676): 345–353.
- Gómez-González, P. J., Campino, S., Phelan, J. E. and Clark, T. G. (2022). Portable sequencing of Mycobacterium tuberculosis for clinical and epidemiological applications. Briefings Bioinf. 23(5): e1093/bib/bbac256.
- Sereika, M., Kirkegaard, R. H., Karst, S. M., Michaelsen, T. Y., Sørensen, E. A., Wollenberg, R. D. and Albertsen, M. (2022). Oxford Nanopore R10.4 long-read sequencing enables the generation of near-finished bacterial genomes from pure cultures and metagenomes without short-read or reference polishing. Nat Methods. 19(7): 823–826.
- Dippenaar, A., Goossens, S. N., Grobbelaar, M., Oostvogels, S., Cuypers, B., Laukens, K., Meehan, C. J., Warren, R. M. and van Rie, A. (2022). Nanopore Sequencing for Mycobacterium tuberculosis: a Critical Review of the Literature, New Developments, and Future Opportunities. J Clin Microbiol. 60(1): e00646–21.
- Hu, T., Chitnis, N., Monos, D. and Dinh, A. (2021). Next-generation sequencing technologies: An overview. Hum Immunol. 82(11): 801–811.
- Phelan, J. E., O’Sullivan, D. M., Machado, D., Ramos, J., Oppong, Y. E. A., Campino, S., O’Grady, J., McNerney, R., Hibberd, M. L., Viveiros, M., et al. (2019). Integrating informatics tools and portable sequencing technology for rapid detection of resistance to anti-tuberculous drugs. Genome Med. 11(1): 41.
- Tang, C., Wu, L., Li, M., Dai, J., Shi, Y., Wang, Q., Xu, F., Zheng, L., Xiao, X., Cai, J., et al. (2024). High-throughput nanopore targeted sequencing for efficient drug resistance assay of Mycobacterium tuberculosis. Front Microbiol. 15: e1331656.
Article Information
Publication history
Received: Jul 4, 2024
Accepted: Nov 28, 2024
Available online: Dec 15, 2024
Published: Feb 5, 2025
Copyright
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Tang, C., Xu, F., Zheng, X. and Xiang, G. (2025). Identification of Mycobacterium tuberculosis and its Drug Resistance by Targeted Nanopore Sequencing Technology. Bio-protocol 15(3): e5170. DOI: 10.21769/BioProtoc.5170.
Category
Microbiology > Microbial genetics > DNA > DNA sequencing
Molecular Biology > DNA > DNA sequencing
Microbiology > Antimicrobial assay > Antibacterial assay
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









