- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Cochlear Organ Dissection, Immunostaining, and Confocal Imaging in Mice
(*contributed equally to this work) Published: Vol 15, Iss 2, Jan 20, 2025 DOI: 10.21769/BioProtoc.5167 Views: 3773
Reviewed by: Marion HoggMohammed Mostafizur RahmanAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

In vivo Electroporation of Skeletal Muscle Fibers in Mice
Steven J. Foltz [...] Hyojung J. Choo
Jul 5, 2023 1830 Views

Visualization and Analysis of Neuromuscular Junctions Using Immunofluorescence
You-Tian Hsieh and Show-Li Chen
Oct 5, 2024 2617 Views

Isolation and Imaging of Microvessels From Brain Tissue
Josephine K. Buff [...] Sophia M. Shi
Aug 5, 2025 2636 Views
Abstract
The organ of Corti, located in the inner ear, is the primary organ responsible for animal hearing. Each hair cell has a V-shaped or U-shaped hair bundle composed of actin-filled stereocilia and a kinocilium supported by true transport microtubules. Damage to these structures due to noise exposure, drug toxicity, aging, or environmental factors can lead to hearing loss and other disorders. The challenge when examining auditory organs is their location within the bony labyrinth and their small and fragile nature. This protocol describes the dissection procedure for the cochlear organ, followed by confocal imaging of immunostained endogenous and fluorescent proteins. This approach can be used to understand hair cell physiology and the molecular mechanisms required for normal hearing.
Key features
• Protocol for the microdissection of the organ of Corti and suitable preparation for later immunostaining.
• This technique involves the evaluation of mouse cochlea for planar-cell-polarity protein.
• Quantitative and qualitative analysis of hair cell cilia in different dimensions.
Keywords: CochleaGraphical overview
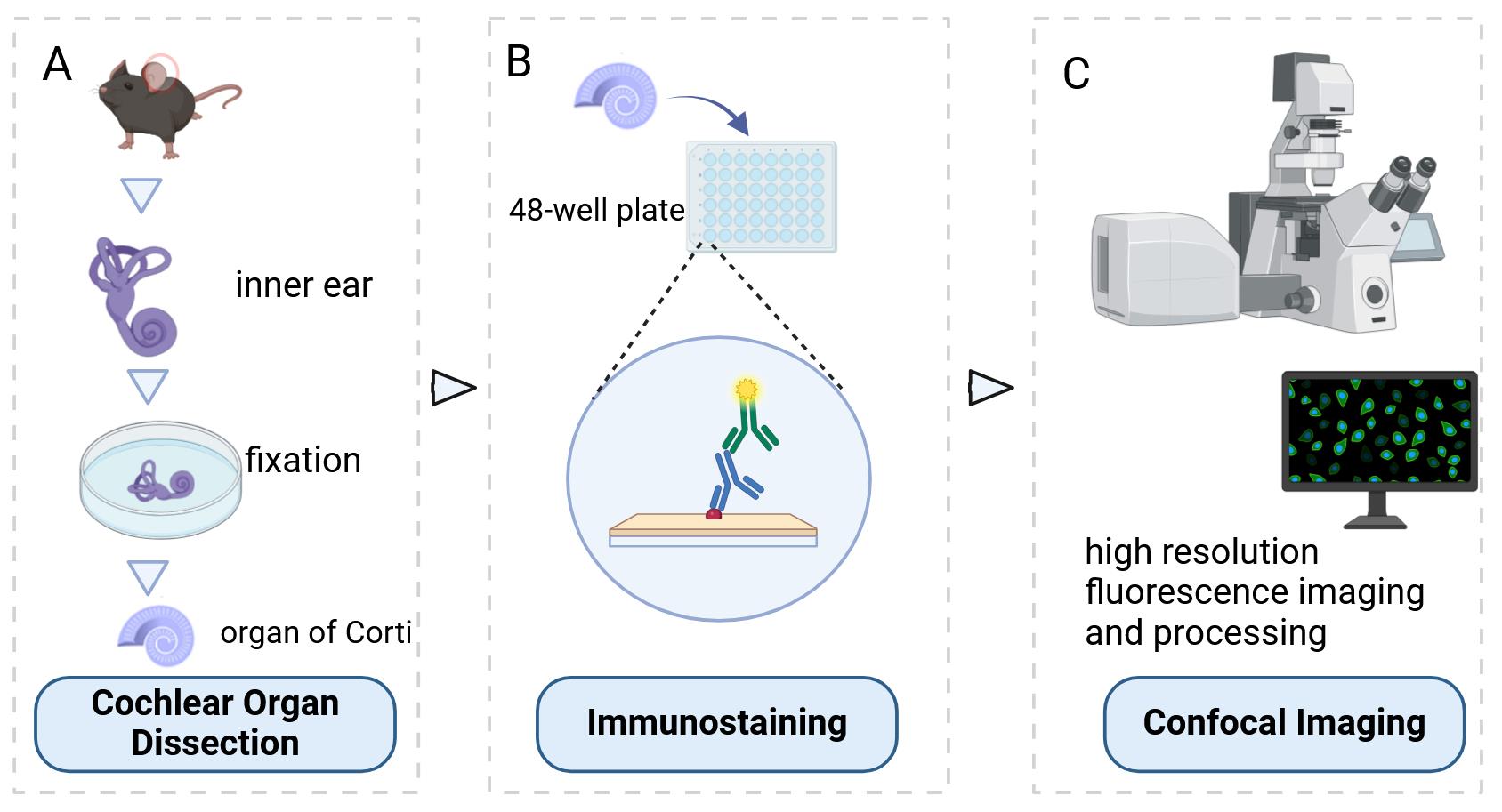
Background
The inner ear, serving as both the auditory organ and local sensory receptor, houses the critical auditory sensor [1,2]. It consists of three rows of outer hair cells and one row of inner hair cells, as well as various supporting cells. These cellular components are meticulously arrayed in a mosaic pattern on the intricate cochlear membrane of the inner ear, demonstrating remarkable flexibility [3–6]. Hair cells convert acoustic signals into electrical signals through the deflection of tip cilia. These cilia are important cellular structures involved in normal morphogenesis and functional activities of hair cells [7,8]. Electrical impulses are ultimately channeled to the cerebral cortex, facilitating auditory perception. Research shows that the primary cause of hearing loss is the involuntary regeneration of damaged hair cells in the mammalian inner ear [9]. The investigation into the auditory impairment mechanism hinges upon the intricate functioning of the Corti organ. Advances in employing specialized immunochemical techniques have significantly enhanced our comprehension of its fundamental operating principles (Figure 1).
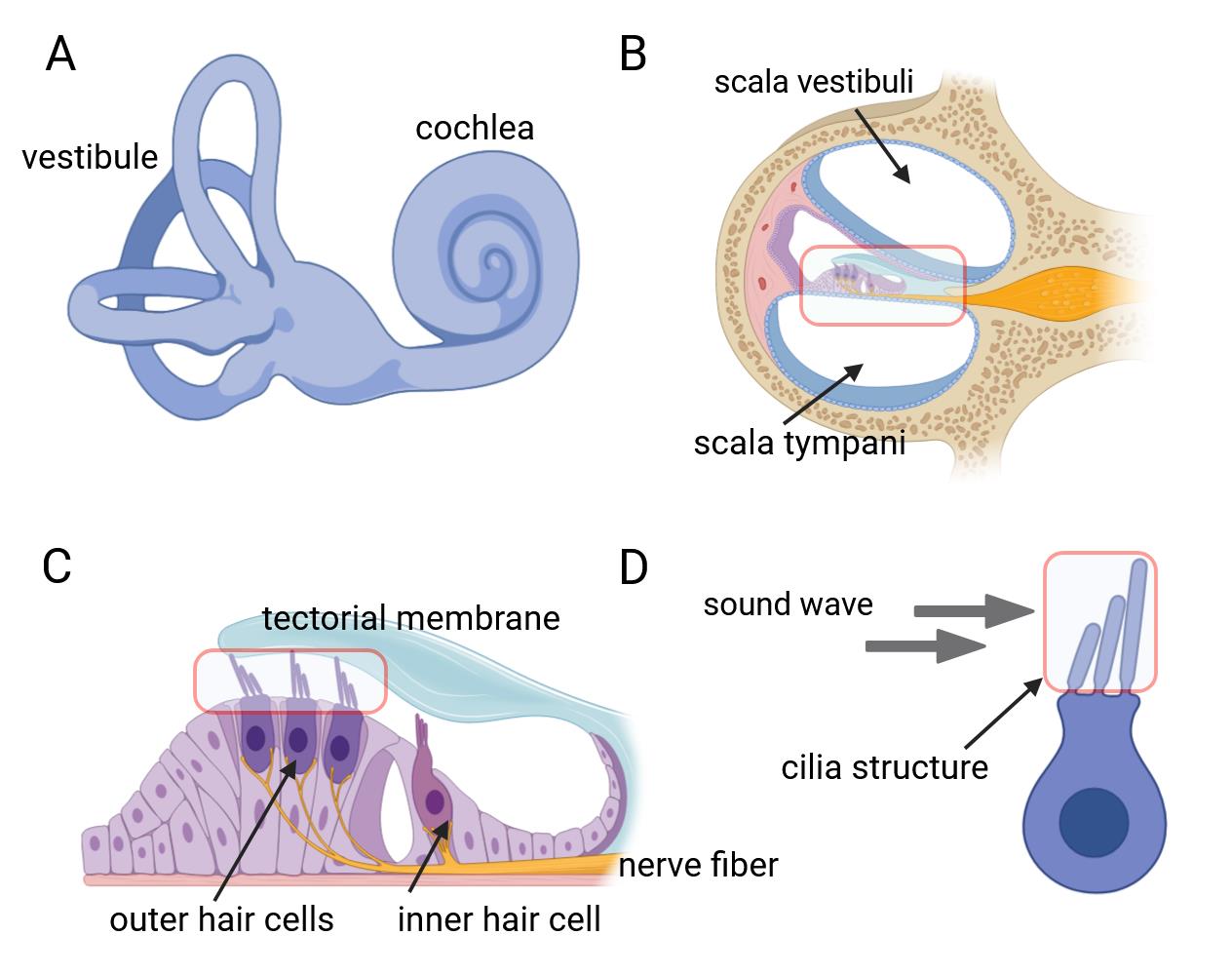
Figure 1. Inner ear anatomy. A, B) The cochlea and a close-up of a single turn cross-section, revealing the organ of Corti. C) The organ of Corti contains mechanosensitive hair cells, which lie beneath the tectorial membrane. D) Hair bundles composed of actin-filled stereocilia arranged in a staircase configuration.
Hair cells exhibit a unique form of planar cell polarity, with their stereocilia bundles oriented toward the peripheral region of the cochlear duct, characterized by a distinct alignment [10]. In this process, several proteins have to be targeted to specific cellular locations to signal the directionality in the cells and to build the specific polarity structure that is unique for the maintenance of the mechanotransduction apparatuses of the sensory hair cells [11–13]. Immunohistochemical techniques are employed to discern hair cell types and precisely locate native proteins contributing to cellular architecture and mechanotransduction capabilities. Furthermore, confocal microscopy enables precise visualization of the ultrastructural features and properties of cochlear lesions, thereby facilitating comprehension of auditory physiological and pathological dynamics. This innovative experimental methodology is extensively employed in fundamental research on auditory sensory cells within the cochlea. We hereby propose a comprehensive protocol designed for the meticulous examination of the mouse auditory organ, encompassing its anatomical structure, immunohistochemical staining procedures, and advanced confocal imaging techniques.
Materials and reagents
Biological materials
1. Rab11a conditional knock-out alleles, Vangl2-Looptail mice (The Jackson Laboratory, catalog number: 000220)
Reagents
1. Phosphate-buffered saline (PBS) (GENOM, catalog number: GNM20012-5)
2. Paraformaldehyde (PFA) (Sigma-Aldrich, catalog number: 158127)
3. Triton X-100 (Sigma, catalog number: 9002-93-1)
4. Normal donkey serum (Merck, Millipore, catalog number: S30)
5. Primary antibodies:
a. Rab11a, 1:200 (Cell Signaling Technology, catalog number: 2413)
b. γ-tubulin, 1:200 (Sigma, catalog number: T6557)
c. Arl13b, 1:1500 (Tamara Caspary, Emory University, Atlanta, GA)
d. Vangl2, 1:200 (R&D Systems, catalog number: AF4815)
e. Fz3, 1:500 (gift from Jeremy Nathans, Johns Hopkins University, Baltimore, MD)
f. LGN, 1:200 (gift from Fumio Matsuzaki, RIKEN)
g. β-Spectrin, 1:200 (BD Transduction Laboratories, catalog number: 612562)
h. MyosinVIIa, 1:200 (Proteus Bioscience Inc, catalog number: 25-6790)
i. Radixin, 1:100 (Abcam, catalog number: ab52495)
j. E-Cadherin, 1:200 (Invitrogen, catalog number: 13-1700)
6. Secondary antibodies:
a. Alexa Fluor® 488 AffiniPure donkey anti-mouse IgG (H+L), 1:1,000 (Jackson Immuno-Research Laboratories, catalog number: 715-545-151)
b. Rhodamine RedTM-X AffiniPure donkey anti-mouse IgG (H+L), 1:1,000 (Jackson Immuno-Research Laboratories, catalog number:715-295-151)
c. Alexa Fluor® 647 AffiniPure donkey anti-mouse IgG (H+L), 1:1,000 (Jackson Immuno-Research Laboratories, catalog number:715-605-151)
7. Phalloidin, 1:1,000 (Sigma-Aldrich, catalog number: P5282)
8. Mounting medium (Vectashield Antifade Medium, catalog number: H-1200)
9. Sodium chloride (NaCl) (Sigma-Aldrich, catalog number: S3014)
10. Potassium chloride (KCl) (Sigma-Aldrich, catalog number: P9541)
11. Sodium phosphate dibasic heptahydrate (NaH2PO4·7H2O) (Sigma-Aldrich, catalog number: S9390)
12. Potassium dihydrogen phosphate (KH2PO4) (Sigma-Aldrich, catalog number: P9791)
Solutions
1. 1× PBS (see Recipes)
2. 4% PFA (see Recipes)
Recipes
1. 1× PBS
| Reagent | Final concentration | Amount |
|---|---|---|
| Sodium chloride | n/a | 8 g |
| Potassium chloride | n/a | 200 mg |
| Sodium phosphate dibasic heptahydrate | n/a | 1.44 g |
| Potassium dihydrogen phosphate | n/a | 240 mg |
| H2O | n/a | 1,000 mL |
| Total | n/a | 1,000 mL |
Adjust pH to 7.4 with HCl and store it at room temperature.
2. 4% PFA
| Reagent | Final concentration | Amount |
|---|---|---|
| Paraformaldehyde (powder) | n/a | 4 g |
| 1× PBS | n/a | 100 mL |
| Total | 4% | 100 mL |
Store paraformaldehyde powder at 4 °C. Heat while stirring under the chemical hood at approximately 60 °C. When the solution is transparent, let it cool down and then filter with Whatman paper to remove undissolved particles. Freeze PFA aliquots in 50 mL tubes at -20 °C. Remember to wear suitable personal protective equipment.
Equipment
1. Dissection microscope (BT, catalog number: BTS-300)
2. Large forceps (Fine Science Tools, catalog number: 11026-15)
3. Fine forceps (Fine Science Tools, catalog number: 11252-00)
4. Very fine forceps (Fine Science Tools, catalog number: 11200-14)
5. Small scissors (Fine Science Tools, catalog number: 91460-11)
6. Confocal microscope (Carl Zeiss Microscopy, model: LSM 510, AxioObserver)
7. 48-well plate (Corning, catalog number: 3548)
8. Petri dish (BD Biosciences, Falcon®, catalog number: 351006)
Software and datasets
1. ImageJ software (http://rsb.info.nih.gov/ij)
Procedure
Below, we describe the step-by-step procedure for cochlear organ dissection, immunostaining, and confocal imaging in mice from E18.5 to P2.
A. Cochlear organ dissection
1. Euthanize the mouse via cervical dislocation or CO2 inhalation (100% CO2).
2. Decapitate the mouse with small scissors. Using large forceps, remove the brain on each side to identify the temporal bones.
3. Under a dissecting microscope, use fine forceps to isolate the bony labyrinths from the temporal bones.
4. Trim away additional tissue and remove the stapes from the oval window.
5. Use the tip of the fine forceps to clear the oval and round windows and make a small hole in the apex of the cochlear spiral. Fix the bony labyrinths in 1.5 mL of 4% PFA for 5 min on ice.
6. Place the bony labyrinths in a dissecting dish containing PBS to facilitate the next step (Figure 2A).
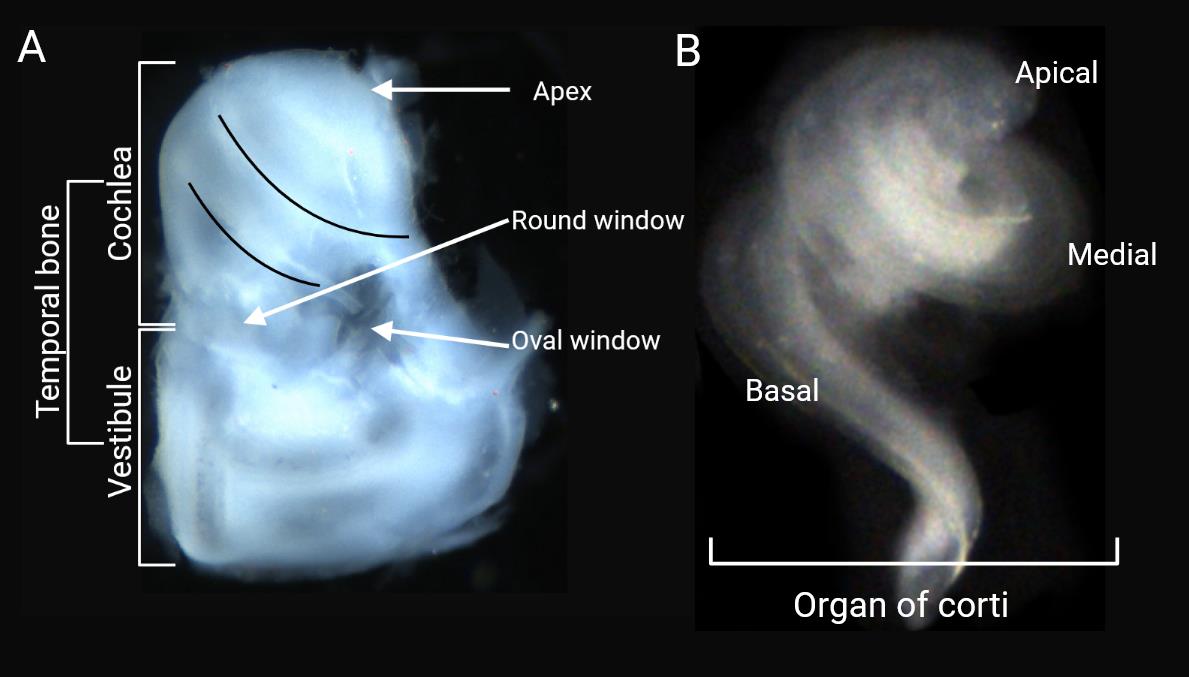
Figure 2. Cochlear organ dissection. A) After the removal of additional tissue from the temporal bone, the otic vesicles are revealed under a light microscope. B) Dissected organ of Corti.
7. Use fine forceps to remove the outer cartilage to expose the cochlear duct.
8. Use fine forceps to slowly remove the stria vascularis. Remove the whole stria in one attempt as far as possible.
9. Use very fine forceps to remove the Reissner’s membrane. Visualize the dorsal aspect of the cochlear duct, including the sensory epithelium.
10. Use very fine forceps to remove the tectorial membrane.
11. Use very fine forceps to remove the intact organ of Corti (Figure 2B, Video 1).
12. Fix the organ of Corti immediately in the 48-well plate containing 0.5 mL of 4% PFA for 2 h at room temperature (RT), 2 h on ice, or overnight at 4 °C.
B. Immunostaining
1. Transfer the organ of Corti to a different well in the 48-well plate containing 0.5 mL of 0.1% Triton X-100 in PBS (PBS-T) for 10–20 min at RT. Agitation is not required for any of these steps. Transfer the organ of Corti with very fine forceps.
2. Place the tissues in 10% normal donkey serum and PBS-T for 1 h at RT.
3. Incubate the organ of Corti tissues in primary antibody in 5% normal donkey serum and PBS-T overnight at 4 °C.
4. Wash with 0.5 mL of PBS-T three times, 1–2 h each, at RT.
5. Incubate the samples in secondary antibodies in 5% normal donkey serum and PBS-T for 2 h at RT or overnight at 4 °C. This procedure should be shielded from light.
6. Wash the sample with PBS-T three times every 1–2 h at RT.
7. Incubate the tissue in phalloidin in 5% normal donkey serum and PBS-T for 30 min at RT.
8. Wash the sample with PBS-T three times every 1–2 h at RT.
C. Confocal imaging
1. Gently transfer the basilar membrane into a drop of PBS placed on a microscope slide.
2. Under the microscope, use very fine forceps to orient the sample facing upward toward the coverslip.
3. Add a drop of mounting medium directly onto the basilar membrane sample. Place one side of the coverslip at an angle against the slide, making contact with the outer edge of the liquid drop, and then lower the cover gently. Take care to avoid air bubbles.
4. Use an adsorbent tissue or a piece of filter paper to remove any excess mounting medium from the edges of the coverslip.
5. Seal the coverslip to the glass slide with nail polish. The sample should be stored at 4 °C in a dark environment until observation.
6. Check the intensity of the labeling and evaluate the background of the experiment by looking at the negative controls.
7. Acquire all pictures in the same conditions (exposure and general settings) for each color channel.
8. Visualize with Zeiss LSM510 confocal microscope equipped with the ZEN software using 20–63× objectives (oil immersion, NA 1.4) and the 488- and 555-nm excitation wavelengths of the laser.
9. Use the Z projection function to provide a two-dimensional view of all the pictures of an image stack by projecting them along the axis perpendicular to the image plane. The Maximum Intensity option creates an image in which each pixel contains the maximum value over all images in the stack.
10. Define specific positions along the cochlear duct, such as 25%, 50%, and 75% from the base, and use these to compare the proteins of interest in cochlear regions.
11. Take pictures using the microscope software for quantification of the particular phenotype as required.
a. Stereocilia morphology: Measure the bundle convexity or the height, defined as the shortest distance between the vertex of the bundle and the line that connects two ends of the bundle. For each genotype, three independent cochleae were counted and quantified, noting the region and hair cell type.
b. Stereocilia orientation: Assess the orientation of each individual bundle relative to a line perpendicular to the planar axis. The angle is defined between the center line and the horizontal line. One line is drawn to bisect the middle of the V-shaped stereocilia bundle, and another line is drawn perpendicular to the planar axis.
c. Cilia quantification: Cilia marked by Arl13b immunostaining are recorded and counted according to different IHC rows and OHC rows. A minimum of 300 IHCs and 1,200 OHCs are counted per region of each genotype. Five individual animal samples are included in the quantification.
12. The above procedures are completed using ImageJ software. The specific steps of the software are as follows (Figure 3):
a. Open the images in ImageJ.
b. Use the straight line tool from the toolbar to connect two ends of the bundle.
c. Use the straight line tool from the toolbar to measure the shortest distance between the vertex of the bundle and the line.
d. Click Analyze > Measure and the length value will appear.
e. Use the angle tool from the toolbar to measure the V-shaped stereocilia bundle.
f. Use the straight line tool from the toolbar to bisect the middle of the V-shaped stereocilia bundle (angle 1 and angle 2 are equal).
g. Use the angle tool from the toolbar to measure the angle between the bisect line and the horizontal line (angle 3).
h. Click Analyze > Measure and the angle value will appear.
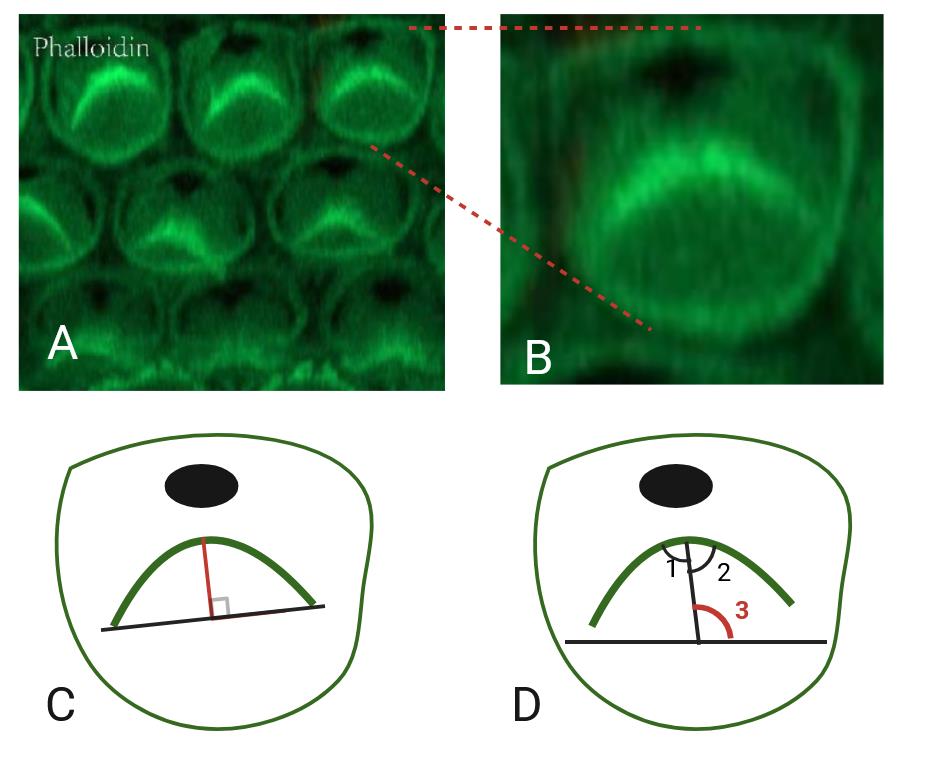
Figure 3. Cochlear hair cells and quantification of the particular aspects. A) Phalloidin staining (green) in the cochlea, showing the stereocilia bundles of hair cells. B) Zoomed-in view of a single hair cell. C) Schematic diagram of length measurement. The red line indicates the length. D) Schematic diagram of angle measurement. One line indicates the bisector of the angle in V-shaped stereocilia bundles (angle 1 and angle 2 are equal). The other line indicates the horizontal line (angle 3, marked in red, indicates the measurement angle).
Validation of protocol
This protocol or parts of it has been used and validated in the following research article:
Knapp et al. [14]. Rab11a Is Essential for the Development and Integrity of the Stereocilia and Kinocilia in the Mammalian Organ of Corti. eNeuro (Figure 1, panels A–D; Figure 4, panels A–E; Figure 5, panels A–H; Figure 6, panels A and B; Figure 7, panels F–I).
General notes and troubleshooting
Troubleshooting
Problem: Unsatisfactory immunostaining results.
Possible cause: Poor quality of samples collected; poor quality of antibodies used; irregular staining procedures.
Solution: Fresh samples should be used as much as possible to prevent problems such as degradation of proteins and severe dehydration. Ensure that the antibody is within its expiration date and use the same batch of antibody as much as possible. Note that it is necessary to avoid light when staining the secondary antibody.
Acknowledgments
Funding was provided by the National Natural Science Foundation of China (NSFC, Grants 82271166, 81970880, and 81771017 to D.R., 82201288 to X.Q.) and by the Natural Science Foundation of Shanghai (Grant No. 22ZR1410100 to D.R.). We thank eNeuro Journal for some illustrating images we used in this paper [14].
Competing interests
The authors declare no competing interests.
Ethical considerations
All procedures were approved by the Emory University Institutional Animal Care and Use committee and met the NIH guidelines.
References
- Jang, M. W., Lim, J., Park, M. G., Lee, J. and Lee, C. J. (2022). Active role of glia‐like supporting cells in the organ of Corti: Membrane proteins and their roles in hearing. Glia. 70(10): 1799–1825.
- Bieniussa, L., Jain, I., Bosch Grau, M., Juergens, L., Hagen, R., Janke, C. and Rak, K. (2023). Microtubule and auditory function – an underestimated connection. Semin Cell Dev Biol. 137: 74–86.
- Xia, A., Udagawa, T., Quiñones, P. M., Atkinson, P. J., Applegate, B. E., Cheng, A. G. and Oghalai, J. S. (2022). The impact of targeted ablation of one row of outer hair cells and Deiters’ cells on cochlear amplification. J Neurophysiol. 128(5): 1365–1373.
- Buswinka, C. J., Rosenberg, D. B., Simikyan, R. G., Osgood, R. T., Fernandez, K., Nitta, H., Hayashi, Y., Liberman, L. W., Nguyen, E., Yildiz, E., et al. (2024). Large-scale annotated dataset for cochlear hair cell detection and classification. Sci Data. 11(1): 416.
- Prajapati-DiNubila, M., Benito-Gonzalez, A., Golden, E. J., Zhang, S. and Doetzlhofer, A. (2019). A counter gradient of Activin A and follistatin instructs the timing of hair cell differentiation in the murine cochlea. eLife. 8: e47613.
- Wan, L., Lovett, M., Warchol, M. E. and Stone, J. S. (2020). Vascular endothelial growth factor is required for regeneration of auditory hair cells in the avian inner ear. Hear Res. 385: 107839.
- Liu, Y., Qi, J., Chen, X., Tang, M., Chu, C., Zhu, W., Li, H., Tian, C., Yang, G., Zhong, C., et al. (2019). Critical role of spectrin in hearing development and deafness. Sci Adv. 5(4): eaav7803.
- Dong, S. H., Kim, S. S., Kim, S. H. and Yeo, S. G. (2020). Expression of aquaporins in inner ear disease. Laryngoscope. 130(6): 1532–1539.
- McQuate, A., Knecht, S. and Raible, D. W. (2023). Activity regulates a cell type-specific mitochondrial phenotype in zebrafish lateral line hair cells. eLife. 12: e80468.
- Scheffer, D. I., Zhang, D. S., Shen, J., Indzhykulian, A., Karavitaki, K. D., Xu, Y. J., Wang, Q., Lin, J. C., Chen, Z. Y., Corey, D. P., et al. (2015). XIRP2, an Actin-Binding Protein Essential for Inner Ear Hair-Cell Stereocilia. Cell Rep. 10(11): 1811–1818.
- Yu, D., Deng, D., Chen, B., Sun, H., Lyu, J., Zhao, Y., Chen, P., Wu, H. and Ren, D. (2022). Rack1 regulates cellular patterning and polarity in the mouse cochlea. Exp Cell Res. 421(2): 113387.
- Okamoto, S., Chaya, T., Omori, Y., Kuwahara, R., Kubo, S., Sakaguchi, H. and Furukawa, T. (2017). Ick Ciliary Kinase Is Essential for Planar Cell Polarity Formation in Inner Ear Hair Cells and Hearing Function. J Neurosci. 37(8): 2073–2085.
- Krey, J. F., Chatterjee, P., Dumont, R. A., O’Sullivan, M., Choi, D., Bird, J. E. and Barr-Gillespie, P. G. (2020). Mechanotransduction-Dependent Control of Stereocilia Dimensions and Row Identity in Inner Hair Cells. Curr Biol. 30(3): 442–454.e7.
- Knapp, L., Sun, H., Wang, Y. M., Chen, B. J., Lin, X., Gao, N., Chen, P. and Ren, D. (2023). Rab11a Is Essential for the Development and Integrity of the Stereocilia and Kinocilia in the Mammalian Organ of Corti. eNeuro. 10(6): ENEURO.0420–22.2023.
Article Information
Publication history
Received: Sep 15, 2024
Accepted: Nov 28, 2024
Available online: Dec 10, 2024
Published: Jan 20, 2025
Copyright
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Chen, C., Chen, B., Qian, X., Sun, H., Fu, X. and Ren, D. (2025). Cochlear Organ Dissection, Immunostaining, and Confocal Imaging in Mice. Bio-protocol 15(2): e5167. DOI: 10.21769/BioProtoc.5167.
- Knapp, L., Sun, H., Wang, Y. M., Chen, B. J., Lin, X., Gao, N., Chen, P. and Ren, D. (2023). Rab11a Is Essential for the Development and Integrity of the Stereocilia and Kinocilia in the Mammalian Organ of Corti. eNeuro. 10(6): ENEURO.0420–22.2023.
Category
Cell Biology > Tissue analysis > Tissue isolation
Neuroscience > Basic technology > Tissue dissection
Cell Biology > Cell structure
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










