- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
A Highly Efficient System for Separating Glandular and Non-glandular Trichome of Cucumber Fruit for Transcriptomic and Metabolomic Analysis
(*contributed equally to this work) Published: Vol 15, Iss 1, Jan 5, 2025 DOI: 10.21769/BioProtoc.5154 Views: 1458
Reviewed by: Wenrong He Kailiang BoAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Separation of Microspores from Anthers of Lilium longiflorum (Lily) and Subsequent RNA Extraction
Ming-Che Liu [...] Co-Shine Wang
Feb 20, 2015 10529 Views

Dot Blot Analysis of N6-methyladenosine RNA Modification Levels
Lisha Shen [...] Hao Yu
Jan 5, 2017 29480 Views
Low-cost and High-throughput RNA-seq Library Preparation for Illumina Sequencing from Plant Tissue
Marta Bjornson [...] Pingtao Ding
Oct 20, 2020 8287 Views
Abstract
Cucumber (Cucumis sativus) trichomes play a critical role in resisting external biological and abiotic stresses. Glandular trichomes are particularly significant as they serve as sites for the synthesis and secretion of secondary metabolites, while non-glandular trichomes are pivotal for determining the appearance quality of cucumbers. However, current methods for separating trichomes encounter challenges such as low efficiency and insufficient accuracy, limiting their applicability in multi-omics sequencing studies. This protocol introduces an efficient system designed for the precise separation of glandular and non-glandular trichomes from cucumber fruit. The process begins with the pre-cooling of sorbitol buffer or ethanol solution and the RNA-free treatment of laboratory supplies, followed by sterilization and pre-cooling. After filling glass bottles with pre-cooling buffer and glass beads, cucumber ovaries are then placed in the glass bottles and the trichome is harvested by bead-beating method. The separation process involves sequential filtration through various steel sieves and centrifugation to separate trichomes. The separated trichomes obtained from this method are well-suited for subsequent multi-omics sequencing analyses. This protocol achieved high precision in separating glandular and non-glandular trichomes, significantly enhancing the efficiency of separation and sample collection processes. This advancement not only addresses existing limitations but also facilitates comprehensive studies aimed at exploring the genetic and biochemical diversity present within cucumber trichomes, thereby opening avenues for broader agricultural and biological research applications.
Key features
• Use cucumber fruits on the day of flowering.
• Pre-cooling and RNA-free treatment ensure supply quality and purity.
• Efficiently separate glandular and non-glandular trichomes.
• Trichome samples are suitable for multi-omics sequencing analysis.
Keywords: Cucumber fruitGraphical overview
Background
With the advancement of biotechnology, there has been a dramatic increase in the number of methods used to separate trichomes in order to study their development, biosynthesis, and metabolism within glandular trichomes. Over the years, methods of separating and collecting trichomes have made great progress in the research process [1].
Earlier methods, such as direct tweezing of trichome cells in tobacco (Nicotiana tabacum) [2] and tweezing of individual trichome cells in Arabidopsis thaliana after quick-freezing plant materials with liquid nitrogen [3] proved inefficient and inaccurate. In tomatoes (Solanum lycopersicum), Pasteur pipettes were employed to collect type VI glandular trichomes from stems and leaves [4]. Advances continued with innovations in mint (Mentha spp.), where chemical and physical methods, aided by buffer solutions, enhanced both plant material protection and glandular trichome yield [5]. These techniques have since been adapted and widely adopted across various plant species [6].
Recent years have seen the bead-beating method emerge as a predominant approach for tomato glandular trichome separation in buffered solutions, followed by filtration and density gradient centrifugation to distinguish between different glandular trichome types [7]. Similarly, this method has proven effective for enriching and purifying cannabis (Cannabis sativa) glandular trichomes [8]. Meanwhile, laser microdissection enables the direct separation of individual glandular trichomes, although its use is time-consuming, inefficient, expensive to collect large numbers of trichomes, and not suitable for large sampling requirements [9].
Previous methods, while foundational, have often suffered from limitations in sampling accuracy and efficiency, primarily focusing on glandular trichome separation while neglecting non-glandular trichomes. In addressing these limitations, this protocol focuses on utilizing North China type cucumber fruits on the day of flowering as the primary research materials. Emphasis is placed on optimizing trichome collection and separation methods to ensure adequate sampling and precise separation, encompassing both glandular and non-glandular trichomes. A substantial yield of trichomes was obtained using the bead-beating method, followed by successful separation of glandular and non-glandular trichomes using steel sieves of varying pore sizes. Non-glandular trichome contents were fully extracted via tissue grinding. Standard samples were prepared for subsequent multi-omics sequencing.
This protocol significantly enhances trichome separation efficiency, achieves precise glandular and non-glandular trichome separation, and serves as a valuable reference for trichome separation in other crops and tissues with a certain firmness. This protocol not only refines current methodologies but also underscores the critical role of precise trichome separation in advancing our understanding of plant biology and biotechnology applications.
Materials and reagents
Biological materials
1. North China type cucumber fruits on the day of flowering (Figure 1)

Figure 1. North China type cucumber fruit on the day of flowering
Reagents
1. Sorbitol (Sigma-Aldrich, CAS number: 50-70-4)
2. Tris-HCl (Sigma-Aldrich, CAS number: 1185-53-1)
3. Sucrose (Sigma-Aldrich, CAS number: 57-50-1)
4. KCl (Sigma-Aldrich, CAS number: 7447-40-7)
5. MgCl2 (Sigma-Aldrich, CAS number: 7786-30-3)
6. Succinic acid (Sigma-Aldrich, CAS number: 110-15-6)
7. EGTA (Sigma-Aldrich, CAS number: 67-42-5)
8. K2HPO4 (Sigma-Aldrich, CAS number: 7758-11-4)
9. Triton X-100 (Sigma-Aldrich, CAS number: 9036-19-5)
10. Anhydrous ethanol (Sigma-Aldrich, CAS number: 64-17-5)
11. RNase remover I (Huayueyang Biotechnology, catalog number: 0416-100)
Solutions
1. Sorbitol buffer (see Recipes)
2. 70% (v/v) ethanol (see Recipes)
Recipes
1. Sorbitol buffer
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Sorbitol | 200 mM | 18.217 g |
| Tris-HCl | 50 mM | 25 mL |
| Sucrose | 20 mM | 3.4229 g |
| KCl | 10 mM | 0.3725 g |
| MgCl2 | 5 mM | 0.5083 g |
| Succinic acid | 5 mM | 0.2958 g |
| EGTA | 1 mM | 0.1902 g |
| K2HPO4 | 0.5 mM | 0.0435 g |
| Triton X-100 | 0.015% (v/v) | 0.075 mL |
| H2O | n/a | Up to 500 mL |
| Total | n/a | 500 mL |
2. 70% (v/v) ethanol
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Anhydrous ethanol | 70% (v/v) | 350 mL |
| H2O (RNA-free) | n/a | 150 mL |
| Total | n/a | 500 mL |
Laboratory supplies
1. Reagent bottles (Fisher Scientific, catalog number: FB800500)
2. Glass beads (Sangon Biotech, catalog number: A500478)
3. 900 μm steel sieves (Shaoxing Shangyu Shengchao Instrument Equipment, catalog number: 20 Mu)
4. 60 μm steel sieves (Shaoxing Shangyu Shengchao Instrument Equipment, catalog number: 250 Mu)
5. 45 μm steel sieves (Shaoxing Shangyu Shengchao Instrument Equipment, catalog number: 325 Mu)
6. Tissue grinder (Sangon Biotech, catalog number: F519062)
7. 50 mL centrifuge tubes (Sangon Biotech, catalog number: F607788)
8. Petri dishes (Sigma-Aldrich, catalog number: BR455751)
Equipment
1. High-speed refrigerated centrifuge (Bioridge, model: TGL-18M)
2. Optical microscope (OLYMPUS, model: CX23)
Procedure
A. Buffer pre-cooling
1. If the samples are obtained for RNA extraction, prepare the 70% (v/v) ethanol solution in advance and pre-cool at -20 °C.
2. If the samples are obtained for metabolite determination, prepare the sorbitol buffer in advance and pre-cool at 4 °C.
B. Pretreatment of laboratory supplies
1. For RNA extraction, perform RNA-free treatment on all experimental supplies involved. Dilute RNase remover I with deionized water at a ratio of 1:1,000. Subsequently, immerse reagent bottles, glass beads, steel sieves, tissue grinder, 50 mL centrifuge tubes, and Petri dishes in this diluted solution for 30 min. Repeat this procedure once with a fresh RNase remover I dilution.
2. After immersing, wrap the supplies in tin foil and autoclave them at 121 °C for 15 min.
3. Dry the laboratory supplies and pre-cool at -20 °C.
C. Trichome harvest
1. Prepare glass bottles containing 30 g of glass beads and 250 mL of pre-cooling buffer. Samples are obtained for RNA extraction using 70% (v/v) ethanol and for metabolite determination using sorbitol solution.
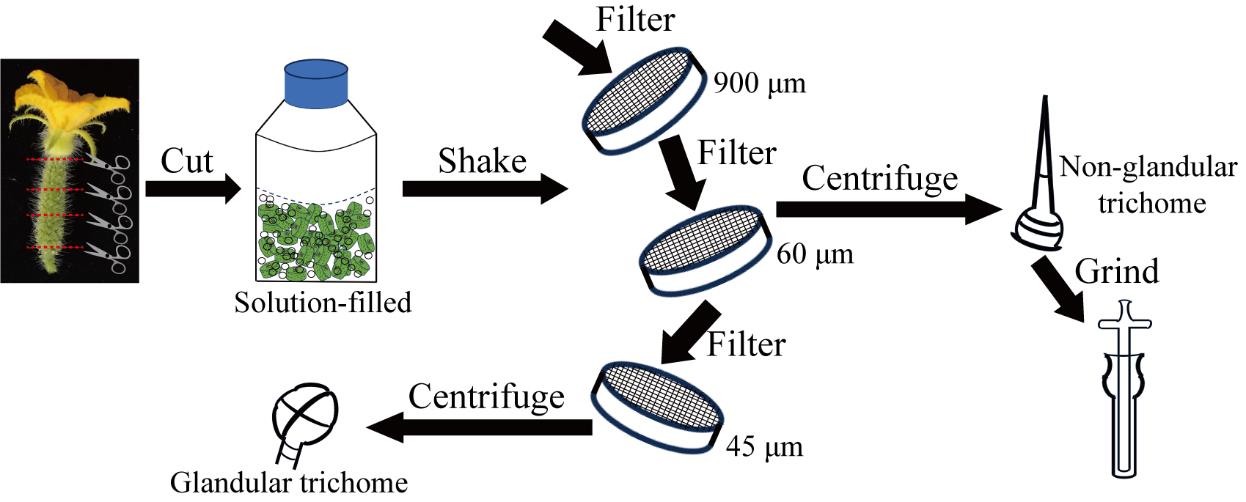
2. On the day of flowering, harvest 25–30 developing ovaries from cucumber plants (Figure 2A). Cut the middle section to a length of approximately 1 cm (Figure 2B) and collect them in glass bottles.
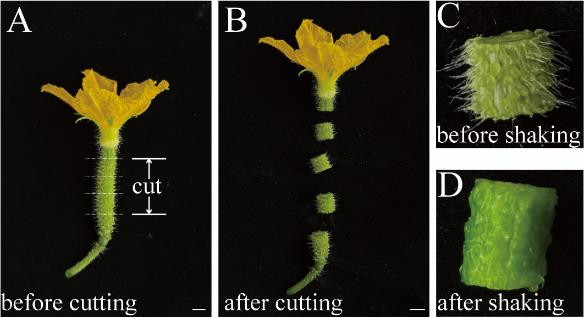
Figure 2. Details of cutting and shaking operation. (A) Developing cucumber ovary on the day of flowering. Bar represents 1 cm. (B) Cucumber ovary after cutting. Bar represents 1 cm. (C) A cut cucumber ovary before shaking. (D) A cut cucumber ovary after shaking.
3. Shake the glass bottles vigorously for 10–15 min until the non-glandular trichomes are almost completely removed from the fruit peels, as visible to the naked eye (Figure 2C and D). Keep the samples at ice-cold temperatures.
D. Separating glandular and non-glandular trichome
1. Remove any remaining cucumber fruit tissues from the glass bottle.
2. Use a 900 μm steel sieve to filter the remaining solution (approximately 250 mL) from the glass bottle in step D1 into a Petri dish and collect the filtrate. Wash the glass bottle with 50 mL of the corresponding solution and collect the additional filtrate. The recovery volume of the filtrate should be higher than 95% of the volume of the added solution.
3. Use a 60 μm steel sieve to filter the filtrate (approximately 285 mL) obtained in step D2 into a new Petri dish and collect the filtrate. Wash the glass bottle with 50 mL of the corresponding solution and collect the additional filtrate. The recovery volume of the filtrate should be higher than 95% of the volume of the added solution.
4. Wash the residual materials on the 60 μm sieve with 50 mL of the corresponding solution and collect them.
5. Observe 20 μL of the recovered products from step D4 under an optical microscope and check that the collected products are non-glandular trichomes. Centrifuge the collected products (approximately 45 mL) in step D4 at 5,000× g for 5 min at 4 °C and remove all supernatants. The precipitate will consist of non-glandular trichomes (approximately 1.5 g).
6. Use a 45 μm steel sieve to filter the filtrate (approximately 320 mL) obtained in step D3 into a new Petri dish and collect the filtrate. Wash the glass bottle with 50 mL of the corresponding solution and collect the additional filtrate. The recovery volume of the filtrate should be higher than 95% of the volume of the added solution.
7. Observe 20 μL of the recovered products from step D6 under an optical microscope and check that the collected products are glandular trichomes. Centrifuge the filtrate (approximately 370 mL) collected in step D6 at 5,000× g for 5 min at 4 °C and remove all supernatants. The precipitate will consist of glandular trichomes (approximately 0.8 g) that are ready for subsequent use.
E. Grinding non-glandular trichome
1. Suspend the non-glandular trichome enrichment products in 800 μL of the corresponding solution and transfer to a tissue grinder. Keep the samples at ice-cold temperatures.
2. Wash the 50 mL centrifuge tube with 500 μL of the corresponding solution and transfer the wash to the tissue grinder. Keep the samples at ice-cold temperatures.
3. Grind the samples with the tissue grinder until no obvious particles remain, then transfer to a new 50 mL centrifuge tube. Keep the samples at ice-cold temperatures.
4. Centrifuge the ground product at 5,000× g for 5 min at 4 °C and remove the supernatant (the main component is the buffer used for extraction). The remaining precipitate consists of non-glandular trichomes ready for RNA extraction. Keep the samples at ice-cold temperatures.
Validation of protocol
This protocol or parts of it has been used and validated in the following research article:
• Feng et al. [10]. Novel players in organogenesis and flavonoid biosynthesis in cucumber glandular trichomes. Plant Physiology (Figures 1–3).
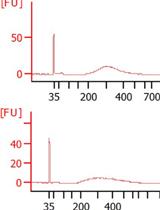
In this paper, we successfully separated glandular and non-glandular trichomes of cucumber fruits. The concentration of total RNAs prepared from glandular and non-glandular trichome samples reached 900 and 60 ng/μL, respectively. We conducted subsequent glandular trichome metabolome sequencing and glandular and non-glandular trichome transcriptome sequencing and mined many genes related to the development and metabolism of cucumber trichomes. The above experimental results prove that our protocol is reliable.
General notes and troubleshooting
General notes
1. RNA sample preparation: Ensure all pretreatments are completed and maintain an ice-cold environment throughout the process.
2. Filtrate collection: The recovery rate of the filtrate at each step should be higher than 95% of the volume of the added solution.
3. Protocol scope: This protocol is designed for obtaining one set of samples. Adjust the sample preparation based on actual requirements.
4. Storage: If the collected products are not used immediately, store them at -80 °C.
Troubleshooting
Problem 1: After shaking for 15 min, many non-glandular trichomes remained on the surface of the fruit peels.
Possible cause: The number of glass beads is too small, or the diameter of glass beads is too small.
Solution: Increase the glass beads to more than 30 g or replace them with glass beads with a diameter of more than 1 mm.
Acknowledgments
This study was supported by the National Key Research and Development Program "Strategic Science and Technology Innovation Cooperation" Key Special Project (2023YFE0206900) and "The 2115 talent development program" of China Agricultural University. A special thanks to the original research paper by Feng et al. [10] (DOI: 10.1093/plphys/kiad236), which described and validated the protocol used in our study, for their invaluable contributions to the field.
Competing interests
There are no conflicts of interest or competing interests.
References
- Feng, Z., Bartholomew, E. S., Liu, Z., Cui, Y., Dong, Y., Li, S., Wu, H., Ren, H. and Liu, X. (2021). Glandular trichomes: new focus on horticultural crops. Hortic Res. 8(1): 158.
- Li, L., Zhao, Y., McCaig, B. C., Wingerd, B. A., Wang, J., Whalon, M. E., Pichersky, E. and Howe, G. A. (2004). The Tomato Homolog of CORONATINE-INSENSITIVE1 Is Required for the Maternal Control of Seed Maturation, Jasmonate-Signaled Defense Responses, and Glandular Trichome Development. Plant Cell. 16(1): 126–143.
- Wienkoop, S., Zoeller, D., Ebert, B., Simon-Rosin, U., Fisahn, J., Glinski, M. and Weckwerth, W. (2004). Cell-specific protein profiling in Arabidopsis thaliana trichomes: identification of trichome-located proteins involved in sulfur metabolism and detoxification. Phytochemistry. 65(11): 1641–1649.
- Xu, J., van Herwijnen, Z. O., Dräger, D. B., Sui, C., Haring, M. A. and Schuurink, R. C. (2018). SlMYC1 Regulates Type VI Glandular Trichome Formation and Terpene Biosynthesis in Tomato Glandular Cells. Plant Cell. 30(12): 2988–3005.
- Gershenzon, J., McCaskill, D., Rajaonarivony, J. I., Mihaliak, C., Karp, F. and Croteau, R. (1992). Isolation of secretory cells from plant glandular trichomes and their use in biosynthetic studies of monoterpenes and other gland products. Anal Biochem. 200(1): 130–138.
- Chen, L., Tian, N., Hu, M., Sandhu, D., Jin, Q., Gu, M., Zhang, X., Peng, Y., Zhang, J. and Chen, Z. (2022). Comparative transcriptome analysis reveals key pathways and genes involved in trichome development in tea plant (Camellia sinensis). Front Plant Sci. 13e997778.
- Bergau, N., Bennewitz, S., Syrowatka, F., Hause, G. and Tissier, A. (2015). The development of type VI glandular trichomes in the cultivated tomato Solanum lycopersicum and a related wild species S. habrochaites. BMC Plant Biol. 15(1): 289.
- Livingston, S. J., Quilichini, T. D., Booth, J. K., Wong, D. C. J., Rensing, K. H., J., Laflamme‐Yonkman, Castellarin, S. D., Bohlmann, J., Page, J. E. and Samuels, A. L. (2020). Cannabis glandular trichomes alter morphology and metabolite content during flower maturation. Plant J. 101(1): 37–56.
- Happyana, N., Agnolet, S., Muntendam, R., Van Dam, A., Schneider, B. and Kayser, O. (2013). Analysis of cannabinoids in laser-microdissected trichomes of medicinal Cannabis sativa using LCMS and cryogenic NMR. Phytochemistry. 8751–59.
- Feng, Z., Sun, L., Dong, M., Fan, S., Shi, K., Qu, Y., Zhu, L., Shi, J., Wang, W. and Liu, Y. (2023). Novel players in organogenesis and flavonoid biosynthesis in cucumber glandular trichomes. Plant Physiol. 192(4): 2723–2736.
Article Information
Publication history
Received: Jul 17, 2024
Accepted: Nov 3, 2024
Available online: Nov 20, 2024
Published: Jan 5, 2025
Copyright
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Sun, L., Feng, Z., Wang, F., Qi, Y., An, M., Yang, L., Feng, M., Wang, M., Ren, H. and Liu, X. (2025). A Highly Efficient System for Separating Glandular and Non-glandular Trichome of Cucumber Fruit for Transcriptomic and Metabolomic Analysis. Bio-protocol 15(1): e5154. DOI: 10.21769/BioProtoc.5154.
- Feng, Z., Sun, L., Dong, M., Fan, S., Shi, K., Qu, Y., Zhu, L., Shi, J., Wang, W. and Liu, Y. (2023). Novel players in organogenesis and flavonoid biosynthesis in cucumber glandular trichomes. Plant Physiol. 192(4): 2723–2736.
Category
Plant Science > Plant biochemistry > RNA > RNA extraction
Molecular Biology > RNA > RNA extraction
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









