- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Isolation of Viral Biofilms From HTLV-1 Chronically Infected T Cells and Integrity Analysis
Published: Vol 14, Iss 24, Dec 20, 2024 DOI: 10.21769/BioProtoc.5152 Views: 1831
Reviewed by: Alka MehraJibin SadasivanSrajan KapoorAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Production, Titration and Imaging of Zika Virus in Mammalian Cells
Wesley Freppel [...] Laurent Chatel-Chaix
Dec 20, 2018 11830 Views

Isolation and CryoTEM of Phages Infecting Bacterial Wine Spoilers
Amel Chaïb [...] Claire Le Marrec
Nov 5, 2020 4419 Views

Production, quantification, and infection of Amazonian Phlebovirus (Bunyaviridae)
Carolina Torturella Rath [...] Ulisses Gazos Lopes
Jul 5, 2021 4051 Views
Abstract
The human T-lymphotropic virus type-1 (HTLV-1) is an oncogenic retrovirus that predominantly spreads through cell-to-cell contact due to the limited infectivity of cell-free viruses. Among various modes of intercellular transmission, HTLV-1 biofilms emerge as adhesive structures, polarized at the cell surface, which encapsulate virions within a protective matrix. This biofilm is supposed to facilitate simultaneous virion delivery during infection. Yet, the molecular and functional intricacies of viral biofilms remain largely unexplored, despite their pivotal role in understanding retroviral pathogenesis. In this study, we optimized a protocol to isolate HTLV-1 biofilms from chronically infected T cells, facilitating their structural and molecular characterization using proteomic and super-resolution microscopy analyses. This protocol involves cultivating HTLV-1 chronically infected T cells at high density to facilitate the natural detachment of viral biofilms into the supernatant. Then, employing successive centrifugations, the cells are separated from the detached biofilms, and these structures are pelleted at medium speed (10,000× g). This method circumvents the need for mechanical, chemical, or enzymatic biofilm detachment, bypasses the use of ultracentrifugation, and enables us to resuspend the biofilms in the appropriate buffer for subsequent analyses such as western blotting or super-resolution microscopy imaging as presented.
Key features
• Isolation of viral biofilms from HTLV-1 chronically infected T cells after 4 days of culture at high cellular density.
• Structural analysis of viral biofilms using super-resolution microscopy techniques.
• Experiments performed in vitro within a confined biosafety level 3 (BSL3) environment.
• This protocol requires at least five days to complete.
Keywords: HTLV-1Graphical overview
Fluorescent HTLV-1 viruses embedded in a biofilm
Background
The human T-lymphotropic virus type-I (HTLV-1) is an oncogenic retrovirus estimated to infect 5–10 million people worldwide [1]. Infected cells are characterized by the integration of the reverse-transcribed viral genome (proviral DNA) in the host genome, which can be expressed using the host transcription machinery, generating new virions and thus leading to chronic infection. Although most people remain asymptomatic following HTLV-1 exposure, chronic infections can lead to aggressive pathologies such as adult T-cell leukemia [2] or progressive inflammatory disorders like HTLV-1-associated myelopathy [3]. In vivo, HTLV-1 is primarily detected in CD4(+) T cells, and its dissemination among individuals occurs through three main routes: mother to child during breastfeeding, sexual contact, or exposure to HTLV-1-infected blood products [4]. Overall, this virus represents a major health issue as no existing strategy allows the efficient protection of exposed individuals. Therefore, fundamental research on HTLV-1 dissemination routes is essential to promote the expansion of pertinent therapeutic approaches.
Unlike most retroviruses, evidence strongly suggests that HTLV-1 cell-free virions are poorly infectious in vivo [5,6]. As such, HTLV-1 relies mostly on intercellular contacts to disseminate within its host. Three cell-to-cell transmission modes have been described: virological synapses, cellular conduits, and viral biofilms, which are of interest here [7]. Viral biofilms are cell-surface aggregates of viruses embedded in extracellular matrix components that accumulate virions at one or several poles of the infected T cells [8]. These structures allow the bulk delivery of infectious particles to target cells and may help to protect HTLV-1 virions from immune attacks [9,10]. While the relative importance of the different cell-to-cell transmission routes is difficult to ascertain in vivo, the removal of biofilms by heparin washes reduces the infectious capacity of HTLV-1-producing cells by 80% in vitro [9]. In addition, biofilms detached from HTLV-1-infected cells are infectious in vitro, as opposed to cell-free virions released in the supernatant of the same cell population [9,11], indicating that the transfer of HTLV-1 biofilms through cell-to-cell contact is the most efficient pathway to infect new cells.
To identify molecular factors involved in HTLV-1 biofilm architecture and explore their impact on viral transmission, we sought to isolate these structures, analyze their molecular composition using mass spectrometry, and confirm by super-resolution STED (stimulated-emission-depletion) microscopy [12]. Here, we will detail the protocol of biofilm isolation that we used in Arone et al. [13] and the techniques employed to validate our protocol (i.e., western blotting and STED microscopy). This protocol is an optimized version of procedures published in Pais-Correia et al. [9] and Alais et al. [11]. In these studies, authors used chemical or mechanical methods, respectively, to detach viral biofilms from HTLV-1 chronically infected T cells and used ultracentrifugation to collect them. While our protocol is also based on cell culture at high density to enrich viral biofilms in the supernatant, it circumvents the need for mechanical or chemical biofilm detachment and bypasses the use of ultracentrifugation. Finally, using immuno-staining coupled with super-resolution STED microscopy techniques [13], we confirm that our isolation protocol preserves the integrity of viral biofilms that contain unaltered virions.
Materials and reagents
Biological materials
HTLV-1 chronically infected T cells (C91-PL) derived from umbilical cord blood cells (CVCL_0197, https://www.cellosaurus.org/CVCL_0197).
pHTLV Gag-YFP plasmid was described previously in Heidecker et al. [14] and is a kind gift from former Dr. Derse’s lab.
Phosphate buffered saline (PBS) pH 7,4 (Gibco, catalog number: 11503387)
Roswell Park Memorial Institute (RPMI) medium (Gibco, catalog number: 11875093)
Opti-MEM reduced serum medium (Gibco, catalog number: 31985062)
Fetal calf serum (FCS) (Sigma-Aldrich, catalog number: F7524)
Penicillin-streptomycin (Sigma-Aldrich, catalog number: P4333)
Trypan blue stain 0.4% (Logos biosystems, catalog number: T13001)
Poly-L-lysine hydrobromide, 5 mg (Sigma-Aldrich, catalog number: P6282)
Bovine serum albumin (BSA) lyophilized powder (Sigma-Aldrich, catalog number: A9418)
32% paraformaldehyde (PFA) without methanol (Thermo Fisher Scientific, catalog number: 047377.9L)
Triton X-100 (Sigma-Aldrich, catalog number: X100)
NaCl 5M solution (Thermo Fisher Scientific, catalog number: AM9760G)
NaCl powder (Sigma-Aldrich, catalog number: S9888)
EDTA 0.5M pH 8.0 (Thermo Fisher Scientific, catalog number: 15575020)
40% acrylamide/bisacrylamide (Euromedex, catalog number: EU0063-C)
Saponin (Sigma-Aldrich, catalog number: SAE0073)
NH4Cl (Sigma-Aldrich, catalog number: A9434)
Methanol (Honeywell, catalog number: 14262-1L)
37% hydrochloric acid (HCl) solution (Honeywell, catalog number: 30721)
Trizma base, crystalline (Sigma-Aldrich, catalog number: T1503)
Tris-glycine 10× buffer (Euromedex, catalog number: EU0550)
Tris-glycine SDS 10× buffer (Euromedex, catalog number: EU0510)
Tris/HCl 1M pH 7.4 (Thermo Fisher Scientific, catalog number: J67501.AE)
10% ammonium persulfate (APS) (Thermo Fisher Scientific, catalog number: 17874)
TEMED (Sigma-Aldrich, catalog number: T9281)
Laemmli SDS 4× buffer (Thermo Fisher Scientific, catalog number: J60015.AC)
Sterile Braun water (Dutscher, catalog number: 921120)
PageRuler prestained protein ladder (Thermo Fisher Scientific, catalog number: 26616)
Tween 20 (Thermo Fisher Scientific, catalog number: 85115)
Milk powder (Régilait)
ECL prime western blotting detection reagent (Amersham, catalog number: 10308449)
Anti-HTLV-1 Gagp19 primary mouse antibody (Zeptometrix, catalog number: 801108)
Anti-HTLV-1 Envgp46 primary mouse antibody (Zeptometrix, catalog number: 0801127)
Anti-YFP primary rabbit antibody (Thermo Fisher Scientific, catalog number: A-11122)
Anti-mouse IgG STAR Red (Abberior, catalog number: 52283)
Anti-rabbit IgG STAR 580 (Abberior, catalog number: 41367)
Anti-mouse IgG HRP (Dako, catalog number: P0260)
TNE buffer (see Recipes)
Protein solubilization buffer (see Recipes)
Poly-L-lysine coating solution (see Recipes)
Fixation buffer (see Recipes)
Permeabilization and saturation buffer (see Recipes)
Tris-HCl 0.5M pH 6.8 (see Recipes)
Tris-HCl 1M pH 8.0 (see Recipes)
Tris-HCl 1.5M pH 8.8 (see Recipes)
Tris-buffered saline (TBS) (see Recipes)
Tris-buffered saline with Tween (TBST) (see Recipes)
10% separating electrophoresis gel (see Recipes)
4% stacking electrophoresis gel (see Recipes)
Western blot running buffer (see Recipes)
Western blot transfer buffer (see Recipes)
TNE buffer
Reagent Final concentration Quantity or Volume Tris/HCl 0.5M pH 7.4 10mM 200 μL NaCl 5M 100mM 400 μL EDTA 0.5M pH 8.0 1mM 40 μL Milli-Q H2O Up to 20 mL Total 20 mL Note: Store at room temperature.
Protein solubilization buffer
Reagent Final concentration Quantity or Volume Triton X-100 0.2% 2 mL PBS pH 7.4 99.8% 8 mL Total 100% 10 mL Note: Store at 4 °C.
Poly-L-lysine coating solution
Reagent Final concentration Quantity or Volume Poly-L-lysine hydrobromide 0.01% 5 mg PBS pH 7.4 99.9% Up to 50 mL Total 100% 50 mL Note: Store at -20 °C as 2 mL aliquots to avoid freeze/thaw cycles.
Fixation buffer
Reagent Final concentration Quantity or Volume 32% paraformaldehyde 4% 10 mL PBS pH 7.4 96% 70 mL Total 100% 80 mL Note: Store at -20 °C as 2 mL aliquots to avoid freeze/thaw cycles.
Permeabilization and saturation buffer
Reagent Final concentration Quantity or Volume Saponin 0.05% 5 mg BSA 3% 1.5 g PBS pH 7.4 96,95% Up to 50 mL Total 100% 50 mL Note: Mix thoroughly to homogenize completely. Use immediately.
Tris-HCl 0.5M pH 6.8
Reagent Final concentration Quantity or Volume Trizma base 30 g HCl (Adjust to pH 6.8) ~ 9.6 mL Milli-Q H2O Up to 500 mL Total 500 mL Note: HCl should be added in increments of 1–5 mL in 400 mL of H2O until the correct pH is reached. Allow the solution to mix completely before adding any more HCl. Complete to 500 mL with H2O. Store at room temperature.
Tris-HCl 1 M pH 8
Reagent Final concentration Quantity or Volume Trizma base 242 g HCl (Adjust to pH 8) ~ 50 mL Milli-Q H2O Up to 2 L Total 2 L Note: HCl should be added in increments of 1–5 mL in 1.6 L of H2O until the correct pH is reached. Allow the solution to mix completely before adding any more HCl. Complete to 500 mL with H2O. Store at room temperature.
Tris-HCl 1.5 M pH 8.8
Reagent Final concentration Quantity or Volume Trizma base 90.75 g HCl (Adjust to pH 8.8) ~ 9 mL Milli-Q H2O Up to 500 mL Total 500 mL Note: HCl should be added in increments of 1–5 mL in 400 mL of H2O until the correct pH is reached. Allow the solution to mix completely before adding any more HCl. Complete to 500 mL with H2O. Store at room temperature.
Tris-buffered saline (TBS) 10×
Reagent Final concentration Quantity or Volume Tris-HCl 1M pH 8 500 mL NaCl powder 7 g Milli-Q H2O Up to 1 L Total 1 L Note: Store at room temperature.
Tris-buffered saline supplemented with Tween (TBST) 1×
Reagent Final concentration Quantity or Volume TBS 10× 1× 100 mL Tween 20 1 mL Milli-Q H2O Up to 1 L Total 1 L Note: Store at room temperature.
10% separating electrophoresis gel
Reagent Final concentration Quantity or Volume 40% acrylamide/bisacrylamide 10% 2.5 mL Tris-HCl 1.5M pH 8.8 2.9 mL 10% APS 100 μL TEMED 10 μL Sterile Braun water 4.5 mL Total 10 mL (1 gel) Note: TEMED and APS should be added last to start gel polymerization. Add 500 μL of isopropanol on top of the gel to straighten it during polymerization. Use immediately.
4% stacking electrophoresis gel
Reagent Final concentration Quantity or Volume 40% acrylamide/bisacrylamide 4% 328 μL Tris-HCl 0.5M pH 6.8 830 μL 10% APS 25 μL TEMED 3.8 µL Milli-Q H2O 2.1 mL Total 3.3 mL (1 gel) Note: Remove the isopropanol covering the 10% gel. TEMED and APS should be added last to start gel polymerization. Use immediately.
Western blot running buffer
Reagent Final concentration Quantity or Volume Tris-glycine SDS 10× 1× 200 mL Milli-Q H2O 1.8 L Total 2 L Note: Store at room temperature.
Western blot transfer buffer
Reagent Final concentration Quantity or Volume Tris-glycine 10× 1× 200 mL Methanol 15% 300 mL Milli-Q H2O 1.5 L Total 2 L Note: Store at 4 °C.
Laboratory supplies
T25 cm3 cell culture flasks (Thermo Fisher Scientific, catalog number: 169900)
6-well plates (Thermo Fisher Scientific, catalog number: 10578911)
5 mL serological pipettes (Gilson, catalog number: F110126)
LUNA cell counting slides (Logos biosystems, catalog number: NC1765657)
1.5 mL sterile Eppendorf safe-lock tubes, microtube (Eppendorf, catalog number: 0030120086)
2 mL sterile Eppendorf safe-lock tubes, microtube (Eppendorf, catalog number: 0030120094)
5 mL sterile open-top thin wall ultra-clear tubes (Beckman Coulter, catalog number: C14279)
50 mL Falcon tubes (Thermo Fisher Scientific, catalog number: 10203001)
Filtered 1,000 μL pipette tips (Thermo Fisher Scientific, catalog number: 11749855)
Filtered 200 μL pipette tips (Thermo Fisher Scientific, catalog number: 11782584)
Filtered 10 μL pipette tips (Thermo Fisher Scientific, catalog number: 94052000)
Gene Pulser/MicroPulser electroporation cuvettes, 0.4 cm gap (Bio-Rad, catalog number: 1652081)
Coated glass bottom fluorodishes (WPI, catalog number: FD35-100)
Polyvinylidene difluoride membranes (Thermo Fisher Scientific, catalog number: 88518)
Electrophoresis gel wrap glass plates set (CBS Scientific, catalog number: MGP100R)
Whatman 3 mm paper for western blot (Cytiva, catalog number: 3630-917)
Equipment
Biosafety cabinet Herasafe KSP Class II (Thermo Fisher Scientific, model: 1300 series A2)
Incubator set to 5% CO2 and 37 °C (Binder, series BD)
ThermoMixer compact (Eppendorf, model: F1.5)
LUNA-II brightfield cell counter (Logos Biosystems, model: C100-Pro)
Eppendorf centrifuge with a fixed-angle rotor for 2 mL Eppendorfs (Thermo Fisher Scientific, model: 5430G)
Optima L-80 XP ultracentrifuge (Beckman Coulter, model: 392051)
FinnPip adjustable volume (P10, P200, P1000) pipettes (Thermo Fisher Scientific, model: monocanal Finnpip)
Pipette controller (Brandtech, model: accu-jet S)
Gene Pulser Xcell electroporation system (Bio-Rad, model: 1652660)
STED super-resolution microscope (Abberior Instruments, model: Expert Line GmbH)
ChemiDoc touch imaging system (Bio-Rad, model: 1708370)
Electrophoresis generator (Apelex, model: PS304 XL)
Double-wide mini-blotter (CBS Scientific, model: EBU-402)
Software and datasets
Image acquisition and analysis
Imspector Image Acquisition & Analysis Software (Abberior, v16.3, 2023)
ImageJ (Fiji, v1.54h, 15/12/2023)
Spectragryph (v1.2.16, 14/07/2022)
GraphPad Prism (v8.3.0, 28/10/2019)
Figure conception and formatting
Inkscape (v1.3.2, 26/11/2023)
Biorender (https://app.biorender.com/)
Procedure
Cell culture and viral biofilm isolation
Cell culture
Note: HTLV-1 chronically infected T cells are cultivated in suspension in a vertical flask (see Figure 1A). The cell suspension (stock culture) is regularly amplified and stored at -150 °C in cryotubes containing fetal calf serum (FCS) supplemented with 10% DMSO. Cells need to be thawed and put in culture at least one week prior to the experiment.
Prepare complete RPMI medium: RPMI 10% FCS, 1% Penicillin/Streptomycin (P/S).
Warm complete RPMI medium and PBS at 37 °C using a water bath.
Pellet HTLV-1 chronically infected T cells from a stock culture at 1,000× g for 5 min.
Resuspend the cells in 10 mL of PBS in a 50 mL falcon tube (wash).
Spin the cell suspension at 1,000× g for 5 min.
Resuspend cells at a concentration of 0.5 × 106 cells/mL in 4 mL of complete RPMI medium.
Transfer the homogenized suspension into a T25 cm3 flask.
Incubate the flask vertically at 37 °C and 5% CO2 for 96 h (Figure 1A).
Note: The doubling time of the C91-PL cell line is 3 days. After 4 days of incubation at high cellular density (0.5 × 106 cells/mL), the cell count reaches approximately 1.25 million cells in 1 mL and therefore a total of 5 million cells in 4 mL.
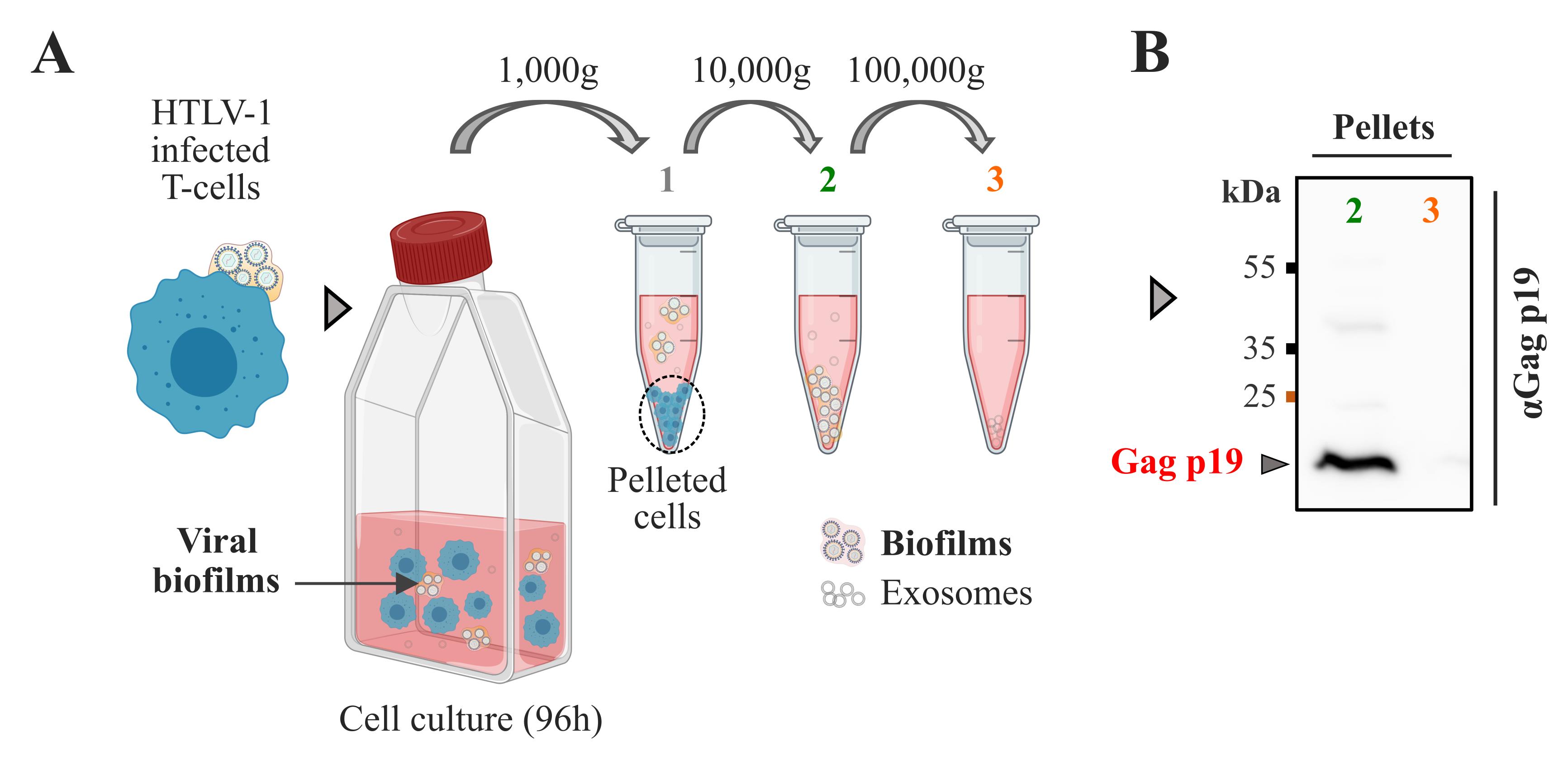
Figure 1. Isolation of HTLV-1 biofilms from chronically infected T cells. A. Protocol for isolation of viral biofilms from chronically infected T cells (C91-PL). C91-PL cells were maintained in culture for 96 h to allow natural biofilm detachment in the supernatant. Step 1: Cells and large cellular debris were removed from the suspension by centrifugation at 1,000× g. Step 2: Biofilm isolation proceeded at 10,000× g. Step 3: Centrifugation at 100,000× g did not pellet more biofilms. B. Immunoblot probed for HTLV-1 Gagp19 showing Gag expression in viral biofilms isolated at 10,000× g (2) or 100,000× g (3).Biofilm isolation using sequential centrifugations
Note: The centrifugation speeds that were tested (10,000× g and 100,000× g) to pellet viral biofilms are described below.
After 96 h of incubation, collect and split the cell suspension (without mixing) in two Eppendorf tubes of 2 mL.
Spin at 1,000× g for 5 min at 4 °C to pellet the cells and clarify the supernatant (Figure 1A.1).
Collect the supernatants (1) and transfer them into two new Eppendorf tubes of 2 mL.
Spin the supernatants (1) at 10,000× g for 40 min at 4 °C (Figure 1A.2).
Collect the supernatants (2) and transfer them into two open-top thin wall ultra-clear tubes of 5 mL.
In parallel, keep the 10,000× g pellets (2) for subsequent analyses.
For western blot, resuspend the pellets in a total volume of 60 μL of PBS with 0.2% Triton.
Incubate for 10 min at room temperature (RT) and store at 4 °C.
Spin the supernatants (2) at 100,000× g for 90 min at 4 °C (Figure 1A.3).
Discard the supernatants and keep the 100,000× g pellets (3) for subsequent analyses.
For the western blot, identical pellets are pooled and resuspended in a total volume of 60 μL of PBS with 0.2% Triton.
Incubate for 10 min at RT and store at 4 °C.
Keep samples at 4 °C for short-term storage (1 week) or -20 °C for long-term storage (several months).
Detection of isolated biofilms using western blot
Note: Prepare a 10% acrylamide/bisacrylamide gel topped by a 4% stacking gel (see Recipes).
All membrane incubation steps are performed under gentle agitation on a lab rocker.
Prepare each sample by mixing in new 1.5 mL Eppendorf tubes:
5 μL of biofilm suspension (10,000× g or 100,000× g fractions)
4 μL of Laemmli SDS 4× buffer
7 μL of Milli-Q H2O
Boil samples at 95 °C for 5 min.
Spin samples at 500× g for 1 min.
Load 4 μL of the prestained protein ladder into the first well of the electrophoresis gel.
Load 16 μL of each sample into the following wells.
Run migration at 80 V until the samples reach the stacking gel.
Run migration at 100 V until the samples reach the end of the running gel.
Pre-wet a PVDF membrane in methanol for 10 min (activation) and briefly rinse in transfer buffer.
Transfer the separated proteins onto the activated PVDF membrane using the wet transfer method at 400 mA for 1 h.
Incubate the membrane in 10 mL of TBST buffer supplemented with 5% milk for 30 min (membrane saturation).
Transfer the membrane in the primary antibody mix containing:
3 mL of 5% milk in TBS-T buffer
3 μL (1:1000) anti-HTLV-1 Gagp19 primary mouse antibody
Incubate overnight at 4 °C under gentle agitation.
Wash the membrane three times in 5% milk in TBS-T buffer for 10 min under gentle agitation.
Transfer the membrane in the secondary antibody mix containing:
10 mL of 5% milk in TBS-T buffer
3.3 μL (1:3,000) anti-mouse HRP-conjugated secondary antibody
Incubate at RT for 2 h under gentle agitation.
Wash the membrane three times in TBST buffer for 10 min under gentle agitation.
Reveal the membrane using the ECL prime kit (Amersham) following the manufacturer’s instructions.
Image the revealed membrane (Figure 1B) with the ChemiDoc imaging system using the following parameters:
Select Chemiluminescence mode.
Set the exposure time manually to 1s.
Note: If your signal is too weak to be detected with 1 s of exposure, select “Rapid Auto-exposure.”
Data analysis (Western blot): HTLV-1 Gagp19 proteins were detected in pellets obtained at 10,000× g (Figure 1B.2) by western blot, demonstrating that viral material has successfully been isolated with this speed. Gagp19 proteins were not detected at 100,000× g by western blot, suggesting that most viral biofilms were pelleted at centrifugation speeds ≤ 10,000× g. Therefore, 10,000× g speed was used to pellet viral biofilms for the following analyses (super-resolution microscopy).
Analysis of isolated biofilms using super-resolution STED microscopy
Note: To confirm the successful isolation of HTLV-1 biofilms, we chose to analyze the viral material pelleted at 10,000× g using stimulated emission-depletion (STED) fluorescence microscopy. This technique allows us to go below 100 nm in x, y resolution and therefore decipher the spatial organization of individual viral particles, measure their diameter, and check if they successfully incorporated viral markers (i.e., Env glycoproteins) (Figure 3).
Electroporation of the pGag-YFP plasmid into HTLV-1 chronically infected T cells
To visualize viral particles, we electroporated HTLV-1 chronically infected T cells with a DNA construct encoding the fluorescent HTLV-1 Gag-YFP protein, which is incorporated into the nascent virions (Figure 2A). This electroporation protocol was optimized for 8 × 106 cells mixed with 10 μg of plasmids.
Warm Opti-MEM (1×) reduced serum medium and PBS at 37 °C using a water bath.
Pellet HTLV-1 chronically infected T cells from a stock culture at 1,000× g for 5 min.
Resuspend cells in 10 mL of PBS in a 50 mL Falcon tube.
Spin the cell suspension at 1,000× g for 5 min (wash).
Count and resuspend cells at a concentration of 2 × 107 cells/mL in 1 mL of Opti-MEM medium.
Transfer 400 μL of this cell suspension (= 8 × 106 cells) into a 1.5 mL Eppendorf tube.
Add 10 μg of pHTLV-1 Gag-YFP plasmids.
Mix gently by pipetting up and down using a P1000 pipette.
Incubate at 37 °C for 30 min.
Mix again gently by pipetting up and down using a P1000 pipette.
Transfer the mix into a 0.4 cm electroporation cuvette.
Place the cuvette into the pod of the Gene Pulser Xcell electroporation system.
Use the following electroporation program:
Voltage: 180 V
Pulse length: 10 ms
Number of pulses: 3
Time between pulses: 1 s
Press the red Pulse button to start the program.
Wait until the electroporation (Figure 2A) is completed (a few seconds).
Transfer 200 μL of the cell suspension into 4 mL of prewarmed RPMI 10% FCS without P/S in a 10 mL Falcon tube.
Mix gently by pipetting up and down using a P1000 pipette.
Divide the cell suspension into two wells of a 6-well plate (volume per well = 2 mL).
Incubate at 37 °C and 5% CO2 for 96 h.
Note: The doubling time of the C91-PL cell line is 3 days. After 4 days of incubation at high cellular density (0.5 × 106 cells/mL), the cell count reaches approximately 1.25 million cells in 1 mL and therefore a total of 5 million cells in 4 mL. 60% of these cells die from the lack of nutrients, which allows spontaneous viral biofilm release.
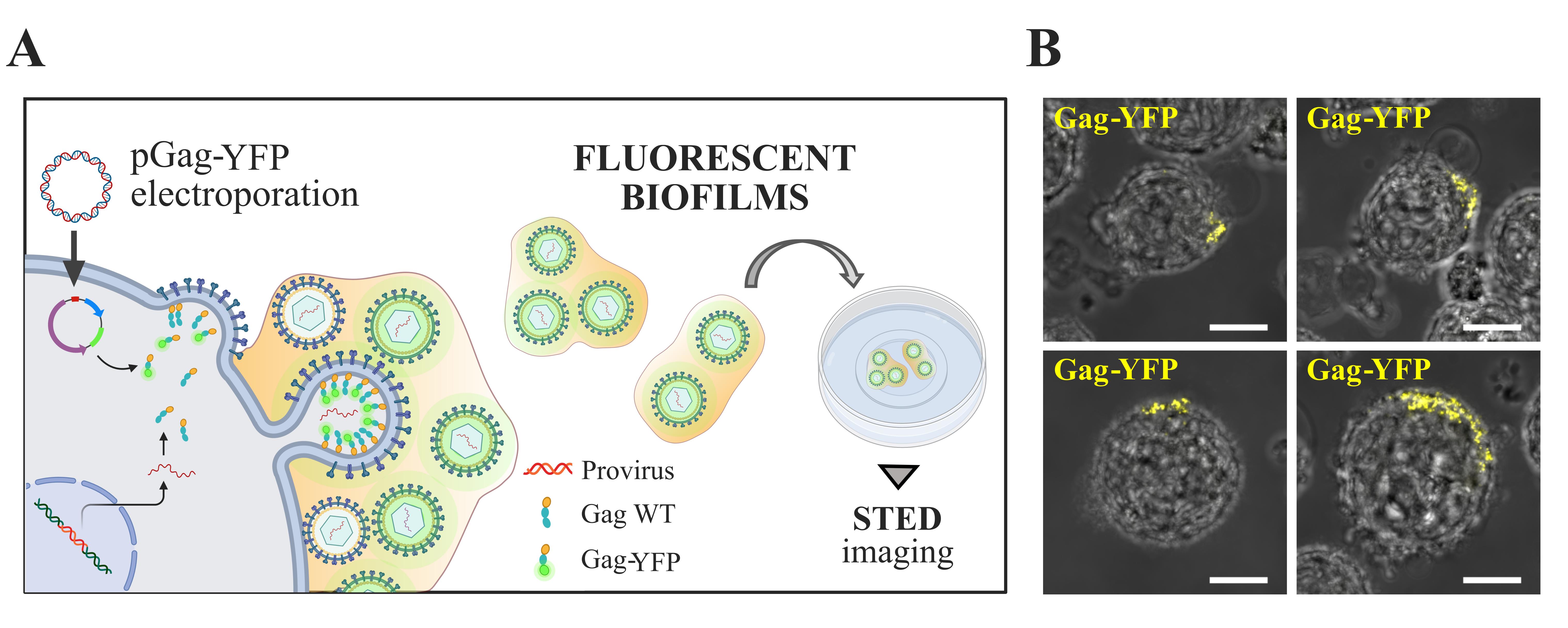
Figure 2. Electroporation of HTLV-1 chronically infected T cells with a plasmid encoding HTLV-1 Gag-YFP fluorescent proteins. A. Scheme: Electroporation of HTLV-1 chronically infected T cells (C91-PL) with a plasmid encoding HTLV-1 Gag-YFP. Gag-YFP(+) biofilms isolated at 10,000× g are then plated on a poly-L-lysine-coated fluorodish for immunofluorescence staining and STED imaging (see below). B. Representative confocal microscopy images of living Gag-YFP(+) chronically infected T cells. Each image is a projection of 10 successive optical z-slices of 1 μm. These images show the successful accumulation of Gag-YFP(+) virions (yellow) at the cell surface 24 h post-electroporation. Scale bars = 10 μm.Remark: We confirmed in Arone et al. [12] that Gag-YFP proteins were successfully incorporated into Gag WT(+) and Env WT(+) virions. This demonstrates that the YFP fluorescent signal reflects wild-type virion accumulation into HTLV-1 biofilms (DOI: 10.1128/mbio.01326-23).
Isolation and plating of Gag-YFP+ biofilms for super-resolution microscopy imaging
After 96 h of incubation, transfer each well into 2 mL Eppendorf tubes (without mixing).
Spin them at 1,000× g for 5 min at 4 °C to pellet the cells (Figure 1A.1) and clarify the supernatant.
Collect the supernatants and transfer them into two new Eppendorf tubes of 2 mL.
Spin the supernatants at 10,000× g for 10 min at 4 °C to pellet viral biofilms as in Figure 1A.2.
Discard the supernatants, resuspend, and pool pellets containing biofilms in a total volume of 60 μL of TNE buffer for storage.
Note: At this point, you can keep the samples at 4 °C for short-term storage (1 week) or -20 °C for long-term storage (several months).
Note: The percentage of cells that dissociates their biofilm is very challenging to estimate. On one hand, even if HTLV-1 chronically infected T cells are releasing biofilms, viral aggregates can still be detected at the cell surface (they are continuously producing viral particles/biofilms), so we cannot discriminate between cells that have “lost their biofilms” and cells that did not. On the other hand, not every cell carries a biofilm at a given timepoint, and these structures are variable in size.
Coat glass-bottom fluorodishes with poly-L-lysine using the following steps:
i. Add 500 μL of PBS containing 0.01% poly-L-lysine in the center of a fluorodish using a P1000 pipette.
ii. Incubate for 30 min at RT.
iii. Remove the poly-L-lysine coating solution.
iv. Wash the bottom of the fluorodish by gently adding and removing 500 μL of PBS.
v. Repeat the washing step three times.
vi. Remove all residual PBS.
Transfer 30 μL of the Gag-YFP(+) biofilms previously isolated into a new 1.5 mL Eppendorf.
Add 470 μL of TNE buffer and gently mix by pipetting up and down to dilute the biofilm suspension.
Transfer the mix (total volume = 500 μL) in the poly-L-lysine coated fluorodish (Figure 2).
Incubate for 30 min at RT in the dark to let the biofilms adhere to the coated glass.
Add 1.5 mL of TNE buffer and proceed to the fixation and staining steps (see below).
Staining of Gag-YFP+ biofilms for stimulated emission-depletion (STED) microscopy:
Note: STED microscopy principle: Using a depletion beam with a “donut-shaped” intensity profile, fluorophore emission can be selectively silenced in the periphery of an excited spot, thus effectively reducing the area of detected fluorescence, and improving the lateral resolution. This allows the detection of individual viral particles in biofilms.
Once biofilms have adhered to the coated glass (Figure 2), remove the TNE buffer.
Add 500 μL of PFA buffer (fixation).
Incubate for 15 min at RT.
Remove PFA and add 500 μL of freshly prepared 50 mM NH4Cl (quenching).
Incubate for 5 min at RT.
Remove NH4Cl and add 1 mL of PBS (wash).
Remove PBS and add 1 mL of PBS containing 0.05% saponin and 3% BSA (permeabilization/saturation buffer).
Incubate for 15 min at RT.
Remove permeabilization/saturation buffer and add the primary antibody mix containing:
i. 200 μL of permeabilization/saturation buffer
ii 1 μL (1:200) anti-YFP primary rabbit antibody
iii. 2 μL (1:100) anti-HTLV-1 Envgp46 primary mouse antibody
Incubate for 90 min at RT.
Remove the primary antibody mix.
Wash the sample by gently adding and removing 1 mL of permeabilization/saturation buffer.
Repeat the washing step three times.
Remove permeabilization/saturation buffer and add the secondary antibody mix containing:
200 μL of permeabilization/saturation buffer
2 μL (1:100) anti-rabbit IgG STAR 580 secondary antibody
2 μL (1:100) anti-mouse IgG STAR Red secondary antibody
Incubate for 90 min at RT in the dark.
Remove the secondary antibody mix.
Wash the sample by gently adding and removing 1 mL of PBS buffer.
Repeat the washing step three times.
Keep the sample in 2 mL of PBS at 4 °C in the dark until STED imaging (Figure 3A).
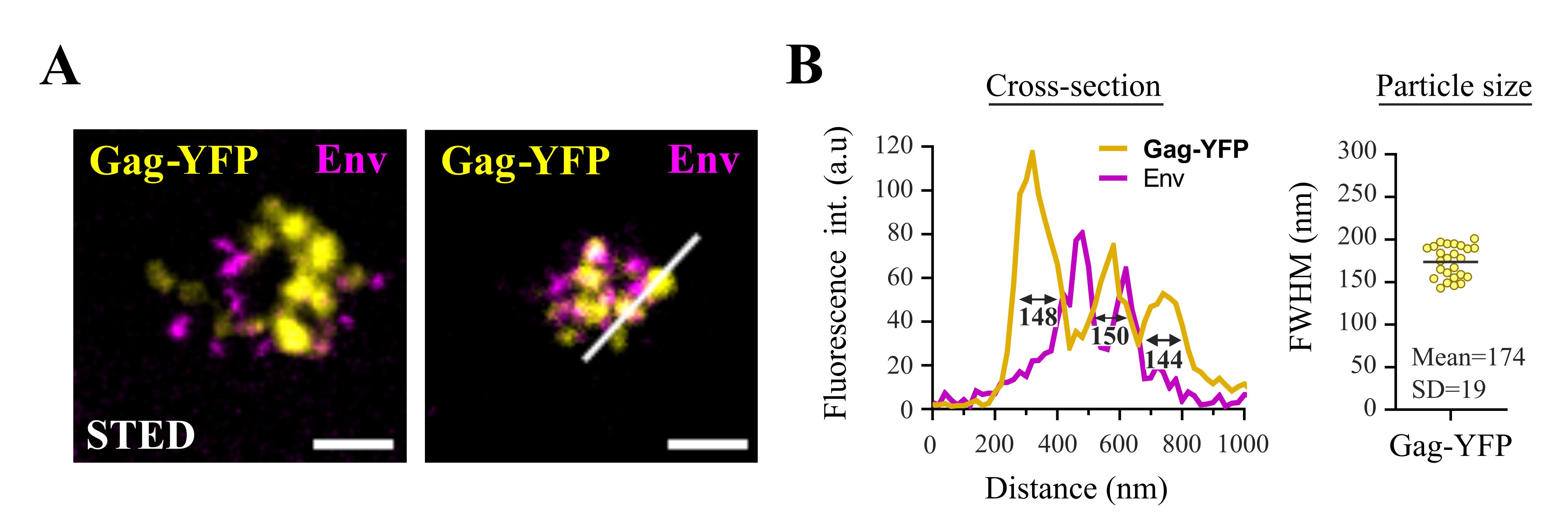
Figure 3. Imaging fluorescent HTLV-1 biofilms using super-resolution STED microscopy. A. Representative STED microscopy images of fixed Gag-YFP(+) biofilms pelleted at 10,000× g and stained for the viral envelope protein (Env). Scale bars = 500 nm. B. The white cross-section, shown in A, is plotted next to the corresponding image (left graph) where the three yellow peaks correspond to individual viral particles (FWHM = 148 nm, 150 nm, and 144 nm). The right plot shows the FWHM distribution of n = 25 cross-sections of Gag-YFP(+) particles and represents the distribution of particles’ diameter. FWHM: full width at half maximum of the Gag-YFP fluorescent signal peak, measured using Spectragryph software. SD: standard deviation of the mean.Imaging parameters (STED): Dual-color STED 2D images (Figure 3B) were acquired on an STED microscope equipped with a 100× oil objective using 580 nm (Star-Orange) and 630 nm (Star-Red) excitation laser sources, coupled with a pulsed 775 nm STED laser. Using 25% of STED laser power, we could obtain a lateral resolution of less than 100 nm. All images were processed with ImageJ software. (DOI: 10.1128/mbio.01326-23.)
Image analysis (STED): Using STED microscopy, we were able to detect Gag-YFP(+) structures in the 10,000× g isolates (Figure 3A). As shown in the representative images in Figure 3A, Gag-YFP(+) signal corresponds to spherical individual particles with a mean diameter (calculated with the full width at half maximum, FWHM, of the fluorescent peak) of 174 nm ± 19 nm (Figure 3B). This size is consistent with the diameter of HTLV-1 virions measured by cryogenic transmission electron microscopy [15]. Moreover, Gag-YFP(+) viral particles successfully incorporated Env glycoproteins (Figure 3A): most Gag-YFP(+) viral cores were in close vicinity with Env signal, which was confirmed in Arone et al. [12]. Although some particles did not exhibit Env glycoproteins at their surface, it is known that HTLV-1 particles contain variable numbers of Env molecules, most of which are unevenly distributed in the viral lipidic envelope [16]. Overall, our results demonstrate the successful isolation of HTLV-1 biofilms containing unaltered Gag(+)/Env(+) viral particles. These isolated biofilms were also analyzed using atomic force microscopy and mass spectrometry by Arone et al. [12].
Validation of protocol
The protocol has been successfully employed in various studies in the lab, as well as in our most recent publication (Arone et al. [12], DOI: 10.1128/mbio.01326-23; Figure 1, Figure 2). In this study, HTLV-1 biofilms were isolated using this protocol and processed by mass spectrometry to do a large-scale identification of biofilm components.
General notes and troubleshooting
In step A.1(f), be sure to resuspend HTLV-1 chronically infected T cells (C91-PL) at a concentration of 0.5 × 106 cells/mL and to incubate the T25 cm3 flask vertically, as high cell density is required for them to survive and grow properly.
In step A.2(a), ensure that cell density/viability has not dropped before harvesting viral biofilms, as it could directly impact the quantity of biofilms collected.
In steps A.2(e, g), pelleted viral biofilms appear as thin white deposits on the bottom of the tubes: try to carefully remove supernatants using a P1000 pipette to avoid losing viral material. Also, remove any residual liquid before resuspending biofilms to avoid any excessive dilution.
In step C1(j), be sure to have a homogenous cell suspension when launching the electroporation step, as they tend to form clusters very rapidly (<10 min). Clusters reduce electroporation efficiency, as the surface area subjected to the electric field decreases when cells are not well dispersed.
In step C1(p), avoid pipetting cellular debris that accumulate at the surface of the suspension after electroporation by plunging the tip at the bottom of the cuvette.
In step C3, always avoid pipetting directly on biofilms during sample preparation for STED imaging, so they are not detached from the glass slides.
Acknowledgments
The authors would like to thank Marie-Pierre Blanchard (MRI, CNRS Montpellier, France) for STED microscopy training and Frederic Eghaian (Abberior, Germany) for providing all STED-compatible secondary antibodies. We also acknowledge the Montpellier Imaging Center for Microscopy (MRI) and the CEMIPAI BSL3 facility for providing excellent working conditions. The illustrations were created with BioRender.com (delphine.muriaux@cemipai.cnrs.fr). This work was supported by the French Agency for Research on AIDS and Viral Hepatitis (grant ANRS0016); institutional funds from the Centre National de la Recherche Scientifique (CNRS); and a 3-year CBS2 Ph.D. fellowship from Montpellier University (UM, France). This protocol was described and validated in the original research paper entitled “HTLV-1 biofilm polarization maintained by tetraspanin CD82 is required for efficient viral transmission” (Arone et al. [12], DOI: 10.1128/mbio.01326-23).
Competing interests
The authors declare no competing interests.
References
- Gessain, A. and Cassar, O. (2012). Epidemiological Aspects and World Distribution of HTLV-1 Infection. Front Microbiol. 3: e00388. https://doi.org/10.3389/fmicb.2012.00388
- Yoshida, M., Seiki, M., Yamaguchi, K. and Takatsuki, K. (1984). Monoclonal integration of human T-cell leukemia provirus in all primary tumors of adult T-cell leukemia suggests causative role of human T-cell leukemia virus in the disease. Proc Natl Acad Sci USA. 81(8): 2534–2537. https://doi.org/10.1073/pnas.81.8.2534
- Nakagawa, M., Izumo, S., Ijichi, S., Kubota, H., Arimura, K., Kawabata, M. and Osame, M. (1995). HTLV-I-associated myelopathy: analysis of 213 patients based on clinical features and laboratory findings. J Neurovirol. 1(1): 50–61. https://doi.org/10.3109/13550289509111010
- Manns, A., Hisada, M. and Grenade, L. L. (1999). Human T-lymphotropic virus type I infection. Lancet. 353(9168): 1951–1958. https://doi.org/10.1016/s0140-6736(98)09460-4
- Donegan, E., Lee, H., Operskalski, E., Shaw, G., Kleinman, S., Busch, M., Stevens, C., Schiff, E., Nowicki, M., Hollingsworth, C., et al. (1994). Transfusion transmission of retroviruses: human T‐lymphotropic virus types I and II compared with human immunodeficiency virus type 1. Transfusion (Paris). 34(6): 478–483. https://doi.org/10.1046/j.1537-2995.1994.34694295061.x
- Miyamoto, K., Tomita, N., Ishii, A., Nishizaki, T., Kitajima, K., Tanaka, T., Nakamura, T., Watanabe, S. and Oda, T. (1984). Transformation of atla‐negative leukocytes by blood components from anti‐ATLA‐positive donors In vitro. Int J Cancer. 33(6): 721–725. https://doi.org/10.1002/ijc.2910330603
- Pique, C. and Jones, K. S. (2012). Pathways of cell-cell transmission of HTLV-1. Front Microbiol. 3: e00378. https://doi.org/10.3389/fmicb.2012.00378
- Maali, Y., Journo, C., Mahieux, R. and Dutartre, H. (2020). Microbial Biofilms: Human T-cell Leukemia Virus Type 1 First in Line for Viral Biofilm but Far Behind Bacterial Biofilms. Front Microbiol. 11: e02041. https://doi.org/10.3389/fmicb.2020.02041
- Pais-Correia, A. M., Sachse, M., Guadagnini, S., Robbiati, V., Lasserre, R., Gessain, A., Gout, O., Alcover, A. and Thoulouze, M. I. (2009). Biofilm-like extracellular viral assemblies mediate HTLV-1 cell-to-cell transmission at virological synapses. Nat Med. 16(1): 83–89. https://doi.org/10.1038/nm.2065
- Thoulouze, M. I. and Alcover, A. (2011). Can viruses form biofilms?. Trends Microbiol. 19(6): 257–262. https://doi.org/10.1016/j.tim.2011.03.002
- Alais, S., Mahieux, R. and Dutartre, H. (2015). Viral Source-Independent High Susceptibility of Dendritic Cells to Human T-Cell Leukemia Virus Type 1 Infection Compared to That of T Lymphocytes. J Virol. 89(20): 10580–10590. https://doi.org/10.1128/jvi.01799-15
- Arone, C., Martial, S., Burlaud-Gaillard, J., Thoulouze, M. I., Roingeard, P., Dutartre, H. and Muriaux, D. (2023). HTLV-1 biofilm polarization maintained by tetraspanin CD82 is required for efficient viral transmission. mBio. 14(6): e01326–23. https://doi.org/10.1128/mbio.01326-23
- Arone, C., Dibsy, R., Inamdar, K., Lyonnais, S., Arhel, N. J., Favard, C. and Muriaux, D. (2021). Illuminating the nanoscopic world of viruses by fluorescence super-resolution microscopy. Virologie. 25(3): 47–60. https://doi.org/10.1684/vir.2021.0908
- Heidecker, G., Lloyd, P. A., Fox, K., Nagashima, K. and Derse, D. (2004). Late Assembly Motifs of Human T-Cell Leukemia Virus Type 1 and Their Relative Roles in Particle Release. J Virol. 78(12): 6636–6648. https://doi.org/10.1128/jvi.78.12.6636-6648.2004
- Maldonado, J., Cao, S., Zhang, W. and Mansky, L. (2016). Distinct Morphology of Human T-Cell Leukemia Virus Type 1-Like Particles. Viruses. 8(5): 132. https://doi.org/10.3390/v8050132
- Cao, S., Maldonado, J. O., Grigsby, I. F., Mansky, L. M. and Zhang, W. (2015). Analysis of Human T-Cell Leukemia Virus Type 1 Particles by Using Cryo-Electron Tomography. J Virol. 89(4): 2430–2435. https://doi.org/10.1128/jvi.02358-14
Article Information
Publication history
Received: May 4, 2024
Accepted: Oct 10, 2024
Available online: Nov 18, 2024
Published: Dec 20, 2024
Copyright
© 2024 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Arone, C., Dutartre, H. and Muriaux, D. (2024). Isolation of Viral Biofilms From HTLV-1 Chronically Infected T Cells and Integrity Analysis. Bio-protocol 14(24): e5152. DOI: 10.21769/BioProtoc.5152.
Category
Immunology > Immune cell staining > Immunodetection
Cell Biology > Cell isolation and culture > Virus isolation
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








